7NTO
 
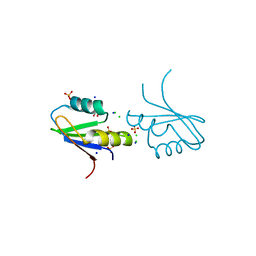 | | The structure of RRM domain of human TRMT2A at 1.23 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Witzenberger, M, Janowski, R, Davydova, E, Niessing, D. | | Deposit date: | 2021-03-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death.
Comput Struct Biotechnol J, 20, 2022
|
|
5JU7
 
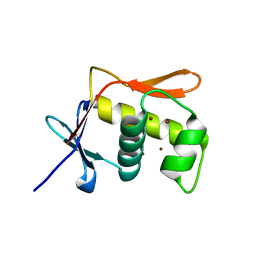 | | DNA BINDING DOMAIN OF E.COLI CADC | | Descriptor: | Transcriptional activator CadC, ZINC ION | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2016-05-10 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator.
Sci Rep, 7, 2017
|
|
6ZSN
 
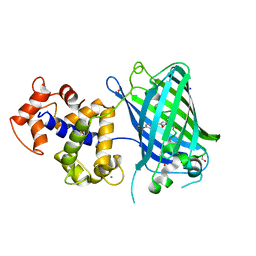 | | Crystal structure of rsGCaMP double mutant Ile80His/Val116Ile in the OFF state (illuminated) | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Green fluorescent protein,Calmodulin, ... | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-07-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 40, 2022
|
|
6ZSM
 
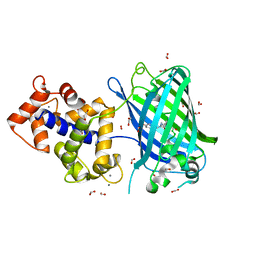 | | Crystal structure of rsGCaMP double mutant Ile80His/Val116Ile in the ON state (non-illuminated) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-07-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 40, 2022
|
|
5L71
 
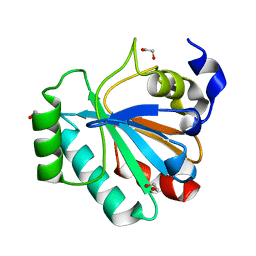 | | Crystal structure of mouse phospholipid hydroperoxide glutathione peroxidase 4 (GPx4) | | Descriptor: | 1,2-ETHANEDIOL, Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | | Authors: | Janowski, R, Scanu, S, Madl, T, Niessing, D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structural studies of mouse phospholipid hydroperoxide glutathione peroxidase 4.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7AUG
 
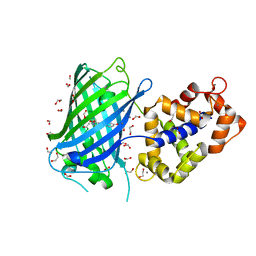 | | Crystal structure of rsGCamP1.3 in the ON state | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-11-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 40, 2022
|
|
5M0H
 
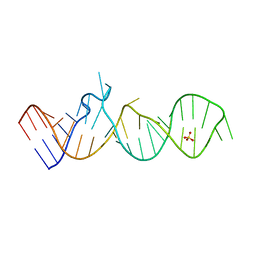 | |
5M0J
 
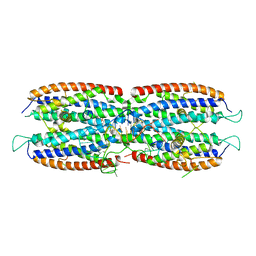 | | Crystal structure of the cytoplasmic complex with She2p, She3p, and the ASH1 mRNA E3-localization element | | Descriptor: | ASH1 E3 (28 nt-loop), MAGNESIUM ION, SWI5-dependent HO expression protein 2,SWI5-dependent HO expression protein 3 | | Authors: | Edelmann, F.T, Janowski, R, Niessing, D. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5CLS
 
 | | Structure of human methionine aminopeptidase-2 complexed with spiroepoxytriazole inhibitor (+)-31a | | Descriptor: | (4R,7R)-7-hydroxy-1-(4-methoxybenzyl)-7-methyl-4,5,6,7-tetrahydro-1H-benzotriazol-4-yl propan-2-ylcarbamate, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Janowski, R, Miller, A.K, Niessing, D. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spiroepoxytriazoles Are Fumagillin-like Irreversible Inhibitors of MetAP2 with Potent Cellular Activity.
Acs Chem.Biol., 11, 2016
|
|
5D6E
 
 | | Structure of human methionine aminopeptidase 2 with covalent spiroepoxytriazole inhibitor (-)-31b | | Descriptor: | (4R,7S)-7-hydroxy-1-(4-methoxybenzyl)-7-methyl-4,5,6,7-tetrahydro-1H-benzotriazol-4-yl propan-2-ylcarbamate, COBALT (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Janowski, R, Miller, A.K, Niessing, D. | | Deposit date: | 2015-08-12 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Spiroepoxytriazoles Are Fumagillin-like Irreversible Inhibitors of MetAP2 with Potent Cellular Activity.
Acs Chem.Biol., 11, 2016
|
|
6FZN
 
 | | SMURFP-Y56R mutant in complex with biliverdin | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, smURFP | | Authors: | Janowski, R, Fuenzalida-Wernera, J.P, Mishra, K, Vetschera, P, Weidenfeld, I, Richter, K, Niessing, D, Ntziachristos, V, Stiel, A.C. | | Deposit date: | 2018-03-15 | | Release date: | 2018-10-17 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a biliverdin-bound phycobiliprotein: Interdependence of oligomerization and chromophorylation.
J. Struct. Biol., 204, 2018
|
|
6FZO
 
 | | SMURFP-Y56F mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Janowski, R, Fuenzalida-Wernera, J.P, Mishra, K, Vetschera, P, Weidenfeld, I, Richter, K, Niessing, D, Ntziachristos, V, Stiel, A.C. | | Deposit date: | 2018-03-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a biliverdin-bound phycobiliprotein: Interdependence of oligomerization and chromophorylation.
J. Struct. Biol., 204, 2018
|
|
6GP9
 
 | |
5D6F
 
 | | Structure of human methionine aminopeptidase-2 complexed with spiroepoxytriazole inhibitor (+)-31b | | Descriptor: | (4S,7R)-7-hydroxy-1-(4-methoxybenzyl)-7-methyl-4,5,6,7-tetrahydro-1H-benzotriazol-4-yl propan-2-ylcarbamate, 1,2-ETHANEDIOL, COBALT (II) ION, ... | | Authors: | Janowski, R, Miller, A.K, Niessing, D. | | Deposit date: | 2015-08-12 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Spiroepoxytriazoles Are Fumagillin-like Irreversible Inhibitors of MetAP2 with Potent Cellular Activity.
Acs Chem.Biol., 11, 2016
|
|
6H0R
 
 | | X-ray structure of SRS2 fragment of Rgs4 3' UTR | | Descriptor: | BARIUM ION, MAGNESIUM ION, SRS2 fragment of Rgs4 3' UTR, ... | | Authors: | Heber, S, Janowski, R, Niessing, D. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Staufen2-mediated RNA recognition and localization requires combinatorial action of multiple domains.
Nat Commun, 10, 2019
|
|
6GQE
 
 | |
5F5F
 
 | | X-ray structure of Roquin ROQ domain in complex with a Selex-derived hexa-loop RNA motif | | Descriptor: | RNA (5'-R(P*UP*GP*AP*CP*UP*GP*CP*GP*UP*UP*UP*UP*AP*GP*GP*AP*GP*UP*UP*A)-3'), Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2015-12-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roquin recognizes a non-canonical hexaloop structure in the 3'-UTR of Ox40.
Nat Commun, 7, 2016
|
|
5F5H
 
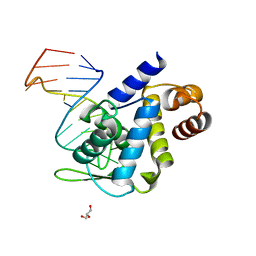 | | X-ray structure of Roquin ROQ domain in complex with Ox40 hexa-loop RNA motif | | Descriptor: | GLYCEROL, RNA (5'-R(P*CP*CP*AP*CP*AP*CP*CP*GP*UP*UP*CP*UP*AP*GP*GP*UP*GP*CP*UP*GP*G)-3'), Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2015-12-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Roquin recognizes a non-canonical hexaloop structure in the 3'-UTR of Ox40.
Nat Commun, 7, 2016
|
|
6Y1C
 
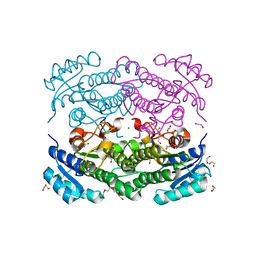 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant D54F | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6Y0Z
 
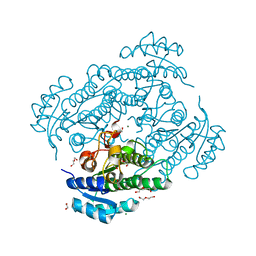 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q126K | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6Y10
 
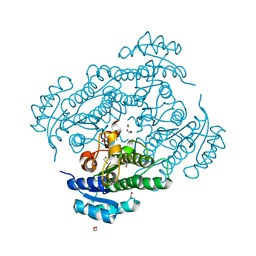 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q126H | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6Y15
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant T102E_Q126K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
6Y1B
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant K32A_Q126K | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
6Y0S
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant T102E | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
6YMC
 
 | | 26-mer stem-loop RNA | | Descriptor: | BARIUM ION, RNA (26-MER) | | Authors: | Janowski, R, Niessing, D. | | Deposit date: | 2020-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple intrinsically disordered RNA-binding motifs cooperate as RNA-folding catalyst and mediate phase transition
To Be Published
|
|
