4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
7YA8
 
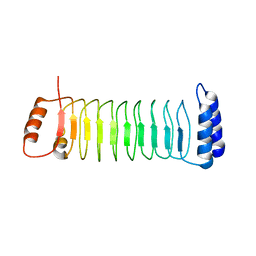 | | The crystal structure of IpaH2.5 LRR domain | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | Deposit date: | 2022-06-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
7YA7
 
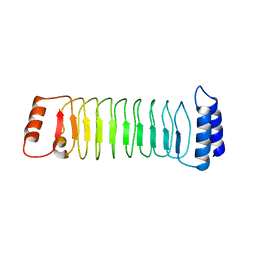 | | The crystal structure of IpaH1.4 LRR domain | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | Deposit date: | 2022-06-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
5D94
 
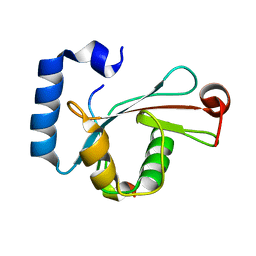 | | Crystal structure of LC3-LIR peptide complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Peptide from FYVE and coiled-coil domain-containing protein 1 | | Authors: | Takagi, K, Mizushima, T, Johansen, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | FYCO1 Contains a C-terminally Extended, LC3A/B-preferring LC3-interacting Region (LIR) Motif Required for Efficient Maturation of Autophagosomes during Basal Autophagy
J.Biol.Chem., 290, 2015
|
|
4YGE
 
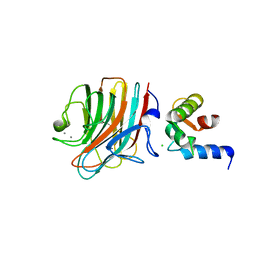 | | Crystal structure of ERGIC-53/MCFD2, trigonal calcium-bound form 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4YGB
 
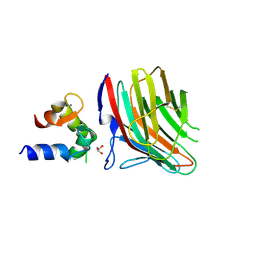 | | Crystal structure of ERGIC-53/MCFD2, monoclinic calcium-free form | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4YGD
 
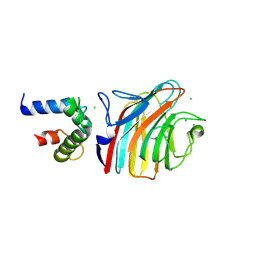 | | Crystal structure of ERGIC-53/MCFD2, monoclinic calcium-bound form 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4YGC
 
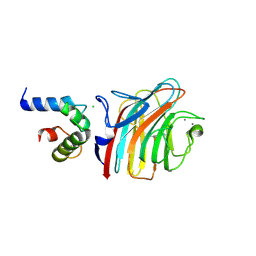 | | Crystal structure of ERGIC-53/MCFD2, monoclinic calcium-bound form 1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7EBC
 
 | | Crystal structure of Isocitrate lyase-1 from Saccaromyces cervisiae | | Descriptor: | Isocitrate lyase, MAGNESIUM ION, TETRAETHYLENE GLYCOL | | Authors: | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
7EBF
 
 | | Cryo-EM structure of Isocitrate lyase-1 from Candida albicans | | Descriptor: | Isocitrate lyase | | Authors: | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
7EBE
 
 | | Crystal structure of Isocitrate lyase-1 from Candida albicans | | Descriptor: | FORMIC ACID, Isocitrate lyase, MAGNESIUM ION | | Authors: | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
5ZI2
 
 | | MDH3 wild type, nad-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Moriyama, S, Nishio, K, Mizushima, T. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5ZI3
 
 | | MDH3 wild type, apo-form | | Descriptor: | GLYCEROL, Malate dehydrogenase | | Authors: | Moriyama, S, Nishio, K, Mizushima, T. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5ZI4
 
 | | MDH3 wild type, nad-oaa-form | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION | | Authors: | Moriyama, S, Nishio, K, Mizushima, T. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1X23
 
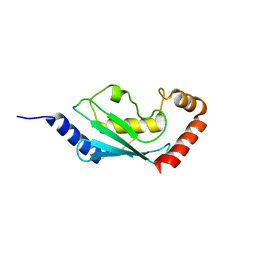 | | Crystal structure of ubch5c | | Descriptor: | Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Nakanishi, M, Teshima, N, Mizushima, T, Murata, S, Tanaka, K, Yamane, T. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of ubch5c
To be Published
|
|
2D1I
 
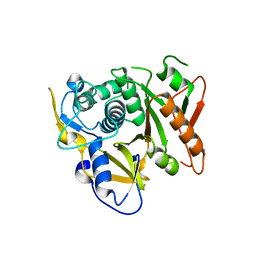 | | Structure of human Atg4b | | Descriptor: | Cysteine protease APG4B | | Authors: | Kumanomidou, T, Mizushima, T, Komatsu, M, Suzuki, A, Tanida, I, Sou, Y.S, Ueno, T, Kominami, E, Tanaka, K, Yamane, T. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Atg4b, a Processing and De-conjugating Enzyme for Autophagosome-forming Modifiers
J.Mol.Biol., 355, 2006
|
|
2DYS
 
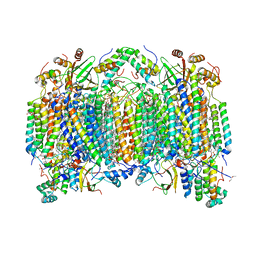 | | Bovine heart cytochrome C oxidase modified by DCCD | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shinzawa-Itoh, K, Aoyama, H, Muramoto, K, Kurauchi, T, Mizushima, T, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2006-09-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase
Embo J., 26, 2007
|
|
2DYR
 
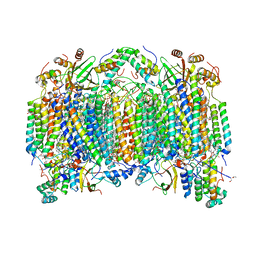 | | Bovine heart cytochrome C oxidase at the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shinzawa-Itoh, K, Aoyama, H, Muramoto, K, Kurauchi, T, Mizushima, T, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2006-09-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase
Embo J., 26, 2007
|
|
2Z5C
 
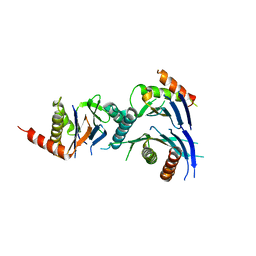 | | Crystal Structure of a Novel Chaperone Complex for Yeast 20S Proteasome Assembly | | Descriptor: | Proteasome component PUP2, Protein YPL144W, Uncharacterized protein YLR021W | | Authors: | Yashiroda, H, Mizushima, T, Okamoto, K, Kameyama, T, Hayashi, H, Kishimoto, T, Kasahara, M, Kurimoto, E, Sakata, E, Suzuki, A, Hirano, Y, Murata, S, Kato, K, Yamane, T, Tanaka, K. | | Deposit date: | 2007-07-03 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes
Nat.Struct.Mol.Biol., 15, 2008
|
|
2ZJD
 
 | | Crystal Structure of LC3-p62 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B precursor, undecameric peptide from Sequestosome-1 | | Authors: | Ichimura, Y, Kumanomidou, T, Sou, Y, Mizushima, T, Ezaki, J, Ueno, T, Kominami, E, Yamane, T, Tanaka, K, Komatsu, M. | | Deposit date: | 2008-03-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Sorting Mechanism of p62 in Selective Autophagy
J.Biol.Chem., 283, 2008
|
|
2Z5B
 
 | | Crystal Structure of a Novel Chaperone Complex for Yeast 20S Proteasome Assembly | | Descriptor: | Protein YPL144W, Uncharacterized protein YLR021W | | Authors: | Yashiroda, H, Mizushima, T, Okamoto, K, Kameyama, T, Hayashi, H, Kishimoto, T, Kasahara, M, Kurimoto, E, Sakata, E, Suzuki, A, Hirano, Y, Murata, S, Kato, K, Yamane, T, Tanaka, K. | | Deposit date: | 2007-07-03 | | Release date: | 2008-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes
Nat.Struct.Mol.Biol., 15, 2008
|
|
3AUL
 
 | | Crystal structure of wild-type Lys48-linked diubiquitin in an open conformation | | Descriptor: | Polyubiquitin-C | | Authors: | Hirano, T, Olivier, S, Yagi, M, Takemoto, E, Hiromoto, T, Satoh, T, Mizushima, T, Kato, K. | | Deposit date: | 2011-02-09 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Conformational dynamics of wild-type Lys48-linked diubiquitin in solution
J.Biol.Chem., 286, 2011
|
|
5B4N
 
 | | Structure analysis of function associated loop mutant of substrate recognition domain of Fbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2 | | Authors: | Nishio, K, Yoshida, Y, Tanaka, K, Mizushima, T. | | Deposit date: | 2016-04-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a function-associated loop mutant of the substrate-recognition domain of Fbs1 ubiquitin ligase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B0N
 
 | | Structure of Shigella effector LRR domain | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Takagi, K, Sasakawa, C, Kim, M, Mizushima, T. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the substrate-recognition domain of the Shigella E3 ligase IpaH9.8
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B0T
 
 | | Structure of Shigella effector LRR domain | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Takagi, K, Sasakawa, C, Kim, M, Mizushima, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the substrate-recognition domain of the Shigella E3 ligase IpaH9.8
Acta Crystallogr.,Sect.F, 72, 2016
|
|
