2C8H
 
 | | Structure of the PN loop Q182A mutant C3bot1 Exoenzyme (NAD-bound state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Properties of Wild-Type and Two Artt Motif Mutants Clostridium Botulinum C3 Exoenzyme Isoform 1 in Different Substrate Complexed States and Crystal Forms.
To be Published
|
|
2CRD
 
 | | ANALYSIS OF SIDE-CHAIN ORGANIZATION ON A REFINED MODEL OF CHARYBDOTOXIN: STRUCTURAL AND FUNCTIONAL IMPLICATIONS | | Descriptor: | CHARYBDOTOXIN | | Authors: | Bontems, F, Roumestand, C, Gilquin, B, Menez, A, Toma, F. | | Deposit date: | 1993-02-17 | | Release date: | 1993-07-15 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Analysis of side-chain organization on a refined model of charybdotoxin: structural and functional implications.
Biochemistry, 31, 1992
|
|
1QUZ
 
 | | Solution structure of the potassium channel scorpion toxin HSTX1 | | Descriptor: | HSTX1 TOXIN | | Authors: | Savarin, P, Romi-Lebrun, R, Zinn-Justin, S, Lebrun, B, Nakajima, T, Gilquin, B, Menez, A. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and functional consequences of the presence of a fourth disulfide bridge in the scorpion short toxins: solution structure of the potassium channel inhibitor HsTX1.
Protein Sci., 8, 1999
|
|
2HEO
 
 | | General Structure-Based Approach to the Design of Protein Ligands: Application to the Design of Kv1.2 Potassium Channel Blockers. | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Z-DNA binding protein 1 | | Authors: | Magis, C, Gasparini, S, Charbonnier, J.B, Stura, E, Le Du, M.H, Menez, A, Cuniasse, P. | | Deposit date: | 2006-06-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based secondary structure-independent approach to design protein ligands: Application to the design of Kv1.2 potassium channel blockers.
J.Am.Chem.Soc., 128, 2006
|
|
1NEA
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A CURAREMIMETIC TOXIN FROM NAJA NIGRICOLLIS VENOM: A PROTON NMR AND MOLECULAR MODELING STUDY | | Descriptor: | TOXIN ALPHA | | Authors: | Zinn-Justin, S, Roumestand, C, Gilquin, B, Bontems, F, Menez, A, Toma, F. | | Deposit date: | 1992-09-22 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a curaremimetic toxin from Naja nigricollis venom: a proton NMR and molecular modeling study.
Biochemistry, 31, 1992
|
|
1BGK
 
 | | SEA ANEMONE TOXIN (BGK) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL, NMR, 15 STRUCTURES | | Descriptor: | BGK | | Authors: | Dauplais, M, Lecoq, A, Song, J, Cotton, J, Jamin, N, Gilquin, B, Roumestand, C, Vita, C, Harvey, A, Menez, A. | | Deposit date: | 1996-05-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures.
J.Biol.Chem., 272, 1997
|
|
1BF0
 
 | | CALCICLUDINE (CAC) FROM GREEN MAMBA DENDROASPIS ANGUSTICEPS, NMR, 15 STRUCTURES | | Descriptor: | CALCICLUDINE | | Authors: | Gilquin, B, Lecoq, A, Desne, F, Guenneugues, M, Zinn-Justin, S, Menez, A. | | Deposit date: | 1998-05-26 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Conformational and functional variability supported by the BPTI fold: solution structure of the Ca2+ channel blocker calcicludine.
Proteins, 34, 1999
|
|
1D5Q
 
 | | SOLUTION STRUCTURE OF A MINI-PROTEIN REPRODUCING THE CORE OF THE CD4 SURFACE INTERACTING WITH THE HIV-1 ENVELOPE GLYCOPROTEIN | | Descriptor: | CHIMERIC MINI-PROTEIN | | Authors: | Vita, C, Drakopoulou, E, Vizzanova, J, Rochette, S, Martin, L, Menez, A, Roumestand, C, Yang, Y.S, Ylisastigui, L, Benjouad, A, Gluckman, J.C. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
1ZEB
 
 | | X-ray structure of alkaline phosphatase from human placenta in complex with 5'-AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1YWH
 
 | | crystal structure of urokinase plasminogen activator receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Llinas, P, Le Du, M.H, Gardsvoll, H, Dano, K, Ploug, M, Gilquin, B, Stura, E.A, Menez, A. | | Deposit date: | 2005-02-18 | | Release date: | 2005-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide
EMBO J., 24, 2005
|
|
2GLQ
 
 | | X-ray structure of human alkaline phosphatase in complex with strontium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, placental type, ... | | Authors: | Llinas, P, Masella, M, Stigbrand, T, Menez, A, Stura, E.A, Le Du, M.H. | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of human alkaline phosphatase in complex with strontium: Implication for its secondary effect in bones.
Protein Sci., 15, 2006
|
|
1ZED
 
 | | Alkaline phosphatase from human placenta in complex with p-nitrophenyl-phosphonate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1ZEF
 
 | | structure of alkaline phosphatase from human placenta in complex with its uncompetitive inhibitor L-Phe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1B45
 
 | | ALPHA-CNIA CONOTOXIN FROM CONUS CONSORS, NMR, 43 STRUCTURES | | Descriptor: | ALPHA-CNIA | | Authors: | Favreau, P, Krimm, I, Le Gall, F, Bobenrieth, M.J, Lamthanh, H, Bouet, F, Servent, D, Molgo, J, Menez, A, Letourneux, Y, Lancelin, J.M. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-09 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Biochemical characterization and nuclear magnetic resonance structure of novel alpha-conotoxins isolated from the venom of Conus consors.
Biochemistry, 38, 1999
|
|
1EW2
 
 | | CRYSTAL STRUCTURE OF A HUMAN PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, PHOSPHATASE, ... | | Authors: | Le Du, M.H, Stigbrand, T, Taussig, M.J, Menez, A, Stura, E.A. | | Deposit date: | 2000-04-21 | | Release date: | 2001-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of alkaline phosphatase from human placenta at 1.8 A resolution. Implication for a substrate specificity.
J.Biol.Chem., 276, 2001
|
|
1KHL
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline Phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHN
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) ZINC FORM | | Descriptor: | Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHJ
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALUMINUM FLUORIDE, Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KCP
 
 | | 3D STRUCTURE OF K-CONOTOXIN PVIIA, A NOVEL POTASSIUM CHANNEL-BLOCKING TOXIN FROM CONE SNAILS, NMR, 22 STRUCTURES | | Descriptor: | KAPPA-CONOTOXIN PVIIA | | Authors: | Savarin, P, Guenneugues, M, Gilquin, B, Lamthanh, H, Gasparini, S, Zinn-Justin, S, Menez, A. | | Deposit date: | 1998-01-27 | | Release date: | 1998-10-14 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of kappa-conotoxin PVIIA, a novel potassium channel-blocking toxin from cone snails.
Biochemistry, 37, 1998
|
|
1KHK
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) | | Descriptor: | Alkaline Phosphatase, MAGNESIUM ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1G1P
 
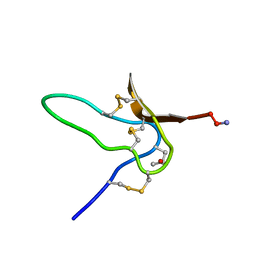 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Barbier, J, Gilles, N, Molgo, J, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-13 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
1G1Z
 
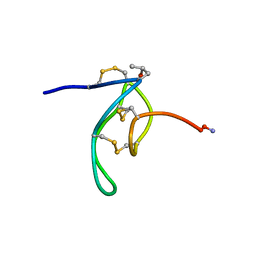 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels, LEU12-PRO13 Cis isomer | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Le Gall, F, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-16 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
2H7Z
 
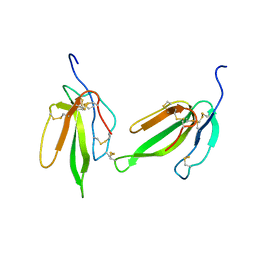 | | Crystal structure of irditoxin | | Descriptor: | Irditoxin subunit A, Irditoxin subunit B | | Authors: | Pawlak, J, Kini, R.M, Stura, E.A, Le Du, M.H. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-29 | | Last modified: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity.
Faseb J., 23, 2009
|
|
3FEV
 
 | | Crystal structure of the chimeric muscarinic toxin MT7 with loop 1 from MT1. | | Descriptor: | Fusion of Muscarinic toxin 1, Muscarinic m1-toxin1, SULFATE ION | | Authors: | Stura, E.A, Menez, R, Mourier, G, Fruchart-Gaillard, C, Menez, A, Servant, D. | | Deposit date: | 2008-12-01 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors.
Plos One, 7, 2012
|
|
