4ZJ7
 
 | |
1FLE
 
 | | CRYSTAL STRUCTURE OF ELAFIN COMPLEXED WITH PORCINE PANCREATIC ELASTASE | | Descriptor: | ELAFIN, ELASTASE | | Authors: | Tsunemi, M, Matsuura, Y, Sakakibara, S, Katsube, Y. | | Deposit date: | 1996-07-04 | | Release date: | 1997-01-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an elastase-specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1.9 A resolution.
Biochemistry, 35, 1996
|
|
5GUA
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138Y mutant | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU9
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138I mutant | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU8
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) wild type | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, SODIUM ION | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4Y65
 
 | |
4Y6I
 
 | |
1WTN
 
 | | The structure of HEW Lysozyme Orthorhombic Crystal Growth under a High Magnetic Field | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Saijo, S, Yamada, Y, Sato, T, Tanaka, N, Matsui, T, Sazaki, G, Nakajima, K, Matsuura, Y. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural consequences of hen egg-white lysozyme orthorhombic crystal growth in a high magnetic field: validation of X-ray diffraction intensity, conformational energy searching and quantitative analysis of B factors and mosaicity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1JDC
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 1) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDA
 
 | | MALTOTETRAOSE-FORMING EXO-AMYLASE | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
3M1I
 
 | |
1JDD
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 2) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
5H2W
 
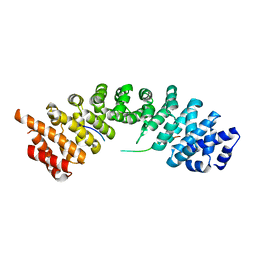 | |
5H2V
 
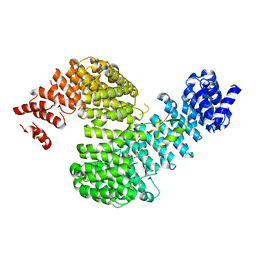 | |
1V8Z
 
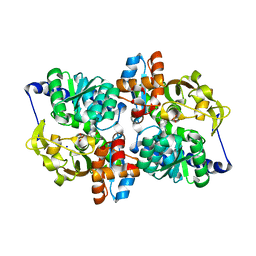 | | X-ray crystal structure of the Tryptophan Synthase b2 Subunit from Hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Hioki, Y, Ogasahara, K, Lee, S.J, Ma, J, Ishida, M, Yamagata, Y, Matsuura, Y, Ota, M, Kuramitsu, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors
Eur.J.Biochem., 271, 2004
|
|
1BF2
 
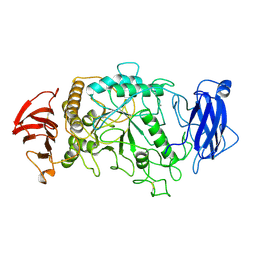 | | STRUCTURE OF PSEUDOMONAS ISOAMYLASE | | Descriptor: | CALCIUM ION, ISOAMYLASE | | Authors: | Katsuya, Y, Mezaki, Y, Kubota, M, Matsuura, Y. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Pseudomonas isoamylase at 2.2 A resolution.
J.Mol.Biol., 281, 1998
|
|
1BFG
 
 | | CRYSTAL STRUCTURE OF BASIC FIBROBLAST GROWTH FACTOR AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Kitagawa, Y, Ago, H, Katsube, Y, Fujishima, A, Matsuura, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of basic fibroblast growth factor at 1.6 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
1WTM
 
 | | X-ray structure of HEW Lysozyme Orthorhombic Crystal formed in the Earth's magnetic field | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Saijo, S, Yamada, Y, Sato, T, Tanaka, N, Matsui, T, Sazaki, G, Nakajima, K, Matsuura, Y. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural consequences of hen egg-white lysozyme orthorhombic crystal growth in a high magnetic field: validation of X-ray diffraction intensity, conformational energy searching and quantitative analysis of B factors and mosaicity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3TPM
 
 | |
3TPQ
 
 | |
3TPO
 
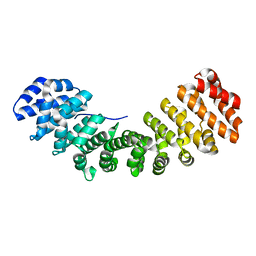 | |
1IPD
 
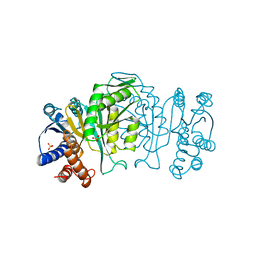 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | Deposit date: | 1992-01-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
5H2X
 
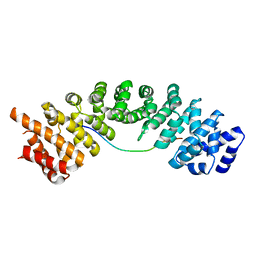 | |
1J0J
 
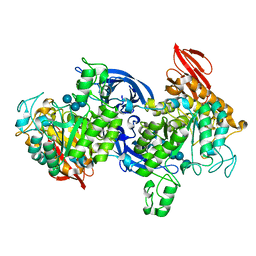 | |
1J0I
 
 | | Crystal structure of neopullulanase complex with panose | | Descriptor: | alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, neopullulanase | | Authors: | Hondoh, H, Kuriki, T, Matsuura, Y. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure and substrate binding of Bacillus stearothermophilus neopullulanase
J.Mol.Biol., 326, 2003
|
|
