7SE7
 
 | | Crystal structure of human Fibrillarin in complex with S-adenosyl-L-methionine | | Descriptor: | S-ADENOSYLMETHIONINE, rRNA 2'-O-methyltransferase fibrillarin | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
7SED
 
 | | Crystal structure of human Fibrillarin in complex with compound 2a | | Descriptor: | FORMIC ACID, N-(piperidin-4-yl)-6-(trifluoromethyl)pyrimidin-4-amine, rRNA 2'-O-methyltransferase fibrillarin | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
7SEC
 
 | | Crystal structure of human Fibrillarin in complex with compound 1a | | Descriptor: | 2-[(8S)-4-oxo-2-(trifluoromethyl)-4,5-dihydropyrazolo[1,5-a]pyrazin-6-yl]acetamide, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
7SE8
 
 | | Crystal structure of human Fibrillarin in complex with fragment 1 from cocktail soak | | Descriptor: | (5S)-3-methyl-7-(trifluoromethyl)pyrrolo[1,2-a]pyrazin-1(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
7SEB
 
 | | Crystal structure of human Fibrillarin in complex with compound 2 from single soak | | Descriptor: | 6-(trifluoromethyl)pyrimidin-4-amine, FORMIC ACID, rRNA 2'-O-methyltransferase fibrillarin | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
7SE9
 
 | | Crystal structure of human Fibrillarin in complex with compound 1 from single soak | | Descriptor: | (5S)-3-methyl-7-(trifluoromethyl)pyrrolo[1,2-a]pyrazin-1(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Shi, Y, El-Deeb, I.M, Masic, V, Hartley-Tassell, L, Maggioni, A, von Itzstein, M, Ve, T. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cofactor Competitive Inhibitors against the Human Methyltransferase Fibrillarin.
Pharmaceuticals, 15, 2021
|
|
6MDR
 
 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
7UXR
 
 | | Crystal structure of the BtTir TIR domain | | Descriptor: | TIR domain protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Vasquez, E, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXS
 
 | | Crystal structure of the BcThsA SLOG domain in complex with 3'cADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, BcThsA, GLYCEROL, ... | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXT
 
 | | Crystal structure of ligand-free SeThsA | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, USG protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Nanson, J.D, Kobe, B, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
1BWK
 
 | | OLD YELLOW ENZYME (OYE1) MUTANT H191N | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH DEHYDROGENASE 1) | | Authors: | Brown, B.J, Deng, Z, Karplus, P.A, Massey, V. | | Deposit date: | 1998-09-24 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the active site of Old Yellow Enzyme. Role of histidine 191 and asparagine 194.
J.Biol.Chem., 273, 1998
|
|
1BWL
 
 | | OLD YELLOW ENZYME (OYE1) DOUBLE MUTANT H191N:N194H | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH DEHYDROGENASE 1) | | Authors: | Brown, B.J, Deng, Z, Karplus, P.A, Massey, V. | | Deposit date: | 1998-09-24 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | On the active site of Old Yellow Enzyme. Role of histidine 191 and asparagine 194.
J.Biol.Chem., 273, 1998
|
|
2L4O
 
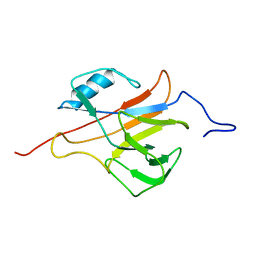 | | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain | | Descriptor: | Cell wall surface anchor family protein | | Authors: | Gentile, M.A, Melchiorre, S, Emolo, C, Moschioni, M, Gianfaldoni, C, Pancotto, L, Ferlenghi, I, Scarselli, M, Pansegrau, W, Veggi, D, Merola, M, Cantini, F, Ruggiero, P, Banci, L, Masignani, V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain: in vivo protection and implications for the molecular mechanisms of pilus polymerization
To be Published
|
|
2N0N
 
 | | NMR solution structure for lactam (5,9) 11mer | | Descriptor: | lactam (5,9) 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N09
 
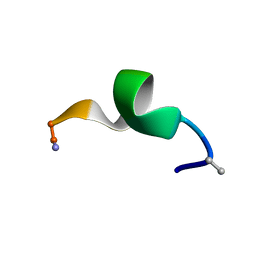 | | NMR structure of a short hydrophobic 11mer peptide in DMSO-d6/H2O (1:3) solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N08
 
 | | NMR structure of a short hydrophobic 11mer peptide in 25 mM SDS solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N0I
 
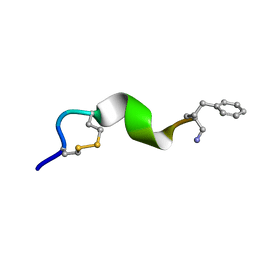 | | NMR solution structure for di-sulfide 11mer peptide | | Descriptor: | di-sulfide 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
1OPO
 
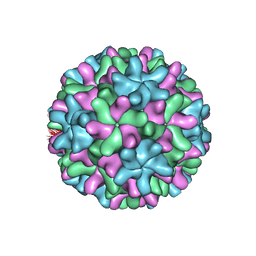 | | THE STRUCTURE OF CARNATION MOTTLE VIRUS | | Descriptor: | CALCIUM ION, Coat protein, SULFATE ION | | Authors: | Morgunova, E, Dauter, Z, Fry, E, Stuart, D, Stel'mashchuk, V, Mikhailov, A.M, Wilson, K.S, Vainshtein, B.K. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The atomic structure of Carnation Mottle Virus capsid protein
Febs Lett., 338, 1994
|
|
2X9W
 
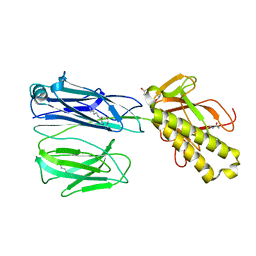 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, GLYCEROL | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9Z
 
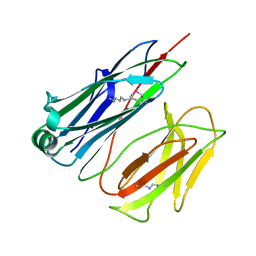 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9Y
 
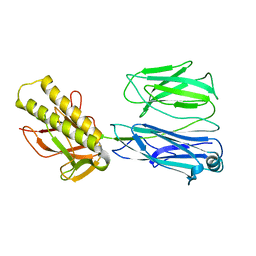 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9X
 
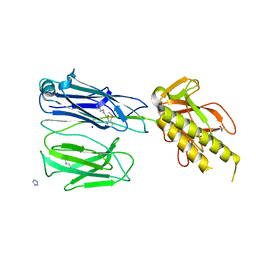 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, IMIDAZOLE, SODIUM ION | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
1DOE
 
 | | THE MOBIL FLAVIN OF 4-OH BENZOATE HYDROXYLASE: MOTION OF A PROSTHETIC GROUP REGULATES CATALYSIS | | Descriptor: | 2,4-DIHYDROXYBENZOIC ACID, BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gatti, D.L, Palfey, B.A, Lah, M.S, Entsch, B, Massey, V, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1994-09-06 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The mobile flavin of 4-OH benzoate hydroxylase.
Science, 266, 1994
|
|
1DOD
 
 | | THE MOBIL FLAVIN OF 4-OH BENZOATE HYDROXYLASE: MOTION OF A PROSTHETIC GROUP REGULATES CATALYSIS | | Descriptor: | 2,4-DIHYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Palfey, B.A, Lah, M.S, Entsch, B, Massey, V, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1994-09-06 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mobile flavin of 4-OH benzoate hydroxylase.
Science, 266, 1994
|
|
1DOC
 
 | | THE MOBIL FLAVIN OF 4-OH BENZOATE HYDROXYLASE: MOTION OF A PROSTHETIC GROUP REGULATES CATALYSIS | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, ... | | Authors: | Gatti, D.L, Palfey, B.A, Lah, M.S, Entsch, B, Massey, V, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1994-09-06 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mobile flavin of 4-OH benzoate hydroxylase.
Science, 266, 1994
|
|
