7UPX
 
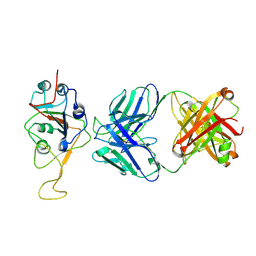 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SP1-77 Fab heavy chain, SP1-77 Fab light chain, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPY
 
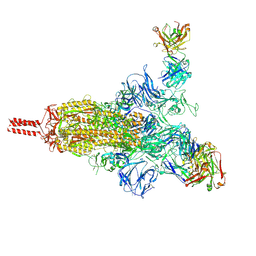 | | An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
3UNM
 
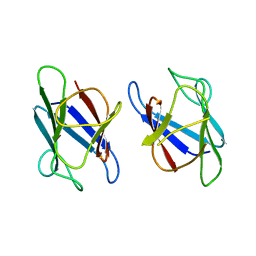 | | Crystal Structure of The Human MDC1 FHA Domain | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain
Nucleic Acids Res., 40, 2012
|
|
3UNN
 
 | |
3UMZ
 
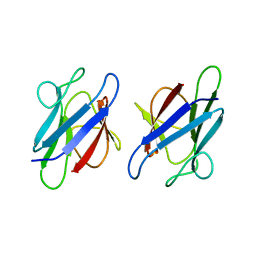 | | Crystal Structure of the human MDC1 FHA Domain | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2011-11-15 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain
Nucleic Acids Res., 40, 2012
|
|
5UC9
 
 | |
5UCA
 
 | |
5UC8
 
 | | Crystal structure of human Heme Oxygenase-2 | | Descriptor: | Heme oxygenase 2 | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Heme Oxygenase 2 Binds Myristate to Regulate Retrovirus Assembly and TLR4 Signaling.
Cell Host Microbe, 21, 2017
|
|
5UXF
 
 | | Crystal Structure of mouse RECON (AKR1C13) in complex with Cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Dihydrodiol dehydrogenase | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-kappa B Activation and Shapes a Proinflammatory Antibacterial State.
Immunity, 46, 2017
|
|
7C7M
 
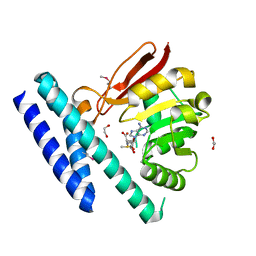 | | The structure of SAM-bound CntL, an aminobutyrate transferase in staphylopine biosysnthesis | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYLMETHIONINE, Staphylopine biosynthesis enzyme CntL | | Authors: | Luo, Z, Luo, S, Zhou, H. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the ligand recognition and catalysis of the key aminobutanoyltransferase CntL in staphylopine biosynthesis.
Faseb J., 35, 2021
|
|
7D5C
 
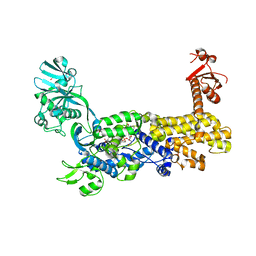 | | IleRS in complex with a tRNA site inhibitor | | Descriptor: | (2E,4S,5S,6E,8E)-10-[(2S,3R,6S,8R,9S)-3-butyl-9-methyl-2-[(1E,3E)-3-methyl-5-oxidanyl-5-oxidanylidene-penta-1,3-dienyl]-3-(4-oxidanyl-4-oxidanylidene-butanoyl)oxy-1,7-dioxaspiro[5.5]undecan-8-yl]-4,8-dimethyl-5-oxidanyl-deca-2,6,8-trienoic acid, 1,2-ETHANEDIOL, Isoleucine--tRNA ligase, ... | | Authors: | Chen, B, Luo, S, Zhou, H. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Inhibitory mechanism of reveromycin A at the tRNA binding site of a class I synthetase.
Nat Commun, 12, 2021
|
|
2ML7
 
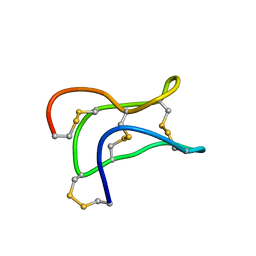 | | Ginsentides: Characterization, Structure and Application of a New Class of Highly Stable Cystine Knot Peptides in Ginseng | | Descriptor: | Specific abundant protein 3 | | Authors: | Wang, S, Nguyen, K, Luo, S, Tam, J, Yang, D. | | Deposit date: | 2014-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ginsentides: Characterization, Structure and Application of a New Class of Highly Stable Cystine Knot Peptides in Ginseng
To be Published
|
|
6L4L
 
 | |
8FSS
 
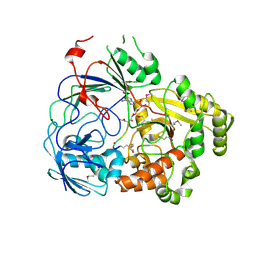 | | Complex Structure of YejA-S481A with Microcin C7 | | Descriptor: | 5'-O-[(R)-amino(3-aminopropoxy)phosphoryl]adenosine, Microcin C7 peptide portion, YejA | | Authors: | Naik, S.K, Dong, S.-H. | | Deposit date: | 2023-01-11 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trojan Horse Peptide Conjugates Remodel the Activity Spectrum of Clinical Antibiotics
To Be Published
|
|
8FSR
 
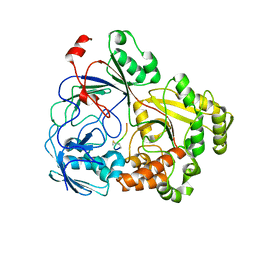 | | Complex Structure of YejA with fMccA | | Descriptor: | Peptide Precursor of Microcin C7, YejA | | Authors: | Naik, S.K, Dong, S.-H. | | Deposit date: | 2023-01-11 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Trojan Horse Peptide Conjugates Remodel the Activity Spectrum of Clinical Antibiotics
To Be Published
|
|
8FSQ
 
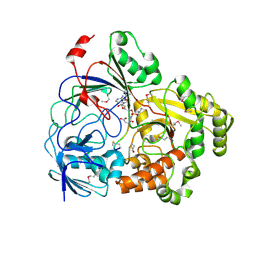 | | Complex Structure of YejA with Microcin C7 | | Descriptor: | 5'-O-[(R)-amino(3-aminopropoxy)phosphoryl]adenosine, Microcin C7 peptide portion, YejA | | Authors: | Naik, S.K, Dong, S.-H. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Trojan Horse Peptide Conjugates Remodel the Activity Spectrum of Clinical Antibiotics
To Be Published
|
|
2MDQ
 
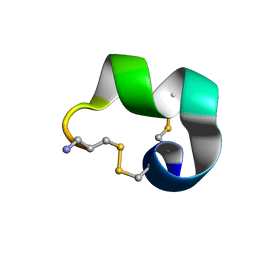 | |
8INK
 
 | | human nuclear pre-60S ribosomal particle - State D | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
8IPX
 
 | |
8IE3
 
 | | human nuclear pre-60S ribosomal particle - State E | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-02-15 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
8IR3
 
 | |
8INE
 
 | |
8IPY
 
 | |
8IDY
 
 | |
