2MHK
 
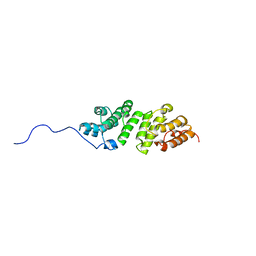 | | E. coli LpoA N-terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Jean, N.L, Bougault, C, Lodge, A, Derouaux, A, Callens, G, Egan, A, Lewis, R.J, Vollmer, W, Simorre, J. | | Deposit date: | 2013-11-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Elongated Structure of the Outer-Membrane Activator of Peptidoglycan Synthesis LpoA: Implications for PBP1A Stimulation.
Structure, 22, 2014
|
|
2VY9
 
 | | Molecular architecture of the stressosome, a signal integration and transduction hub | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST | | Authors: | Marles-Wright, J, Grant, T, Delumeau, O, van Duinen, G, Firbank, S.J, Lewis, P.J, Murray, J.W, Newman, J.A, Quin, M.B, Race, P.R, Rohou, A, Tichelaar, W, van Heel, M, Lewis, R.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Architecture of the "Stressosome," a Signal Integration and Transduction Hub
Science, 322, 2008
|
|
4UG1
 
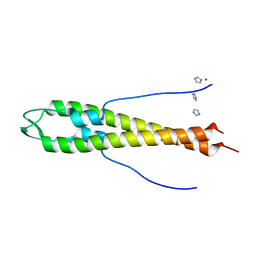 | | GpsB N-terminal domain | | Descriptor: | CELL CYCLE PROTEIN GPSB, IMIDAZOLE, NICKEL (II) ION | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Muller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
4UG3
 
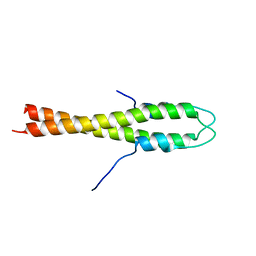 | | B. subtilis GpsB N-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
5FSR
 
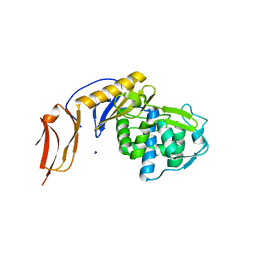 | | Crystal structure of penicillin binding protein 6B from Escherichia coli | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE DACD | | Authors: | Peters, K, Kannan, S, Rao, V.A, Bilboy, J, Vollmer, D, Erickson, S.W, Lewis, R.J, Young, K.D, Vollmer, W. | | Deposit date: | 2016-01-07 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Redundancy of Peptidoglycan Carboxypeptidases Ensures Robust Cell Shape Maintenance in Escherichia Coli
Mbio, 7, 2016
|
|
2EW4
 
 | | Solution structure of MrIA | | Descriptor: | MrIA | | Authors: | Nilsson, K.P, Lovelace, E.S, Caesar, C.E, Tynngard, N, Alewood, P.F, Johansson, H.M, Sharpe, I.A, Lewis, R.J, Daly, N.L, Craik, D.J. | | Deposit date: | 2005-11-02 | | Release date: | 2005-11-15 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Solution structure of chi-conopeptide MrIA, a modulator of the human norepinephrine transporter
Biopolymers, 80, 2005
|
|
2Y8K
 
 | | Structure of CtGH5-CBM6, an arabinoxylan-specific xylanase. | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Firbank, S.J, Correia, M.A, Mazumder, K, Bras, J.L, Zhu, Y, Lewis, R.J, York, W.S, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure and Function of an Arabinoxylan-Specific Xylanase.
J.Biol.Chem., 286, 2011
|
|
2Y0O
 
 | |
2XXQ
 
 | | A widespread family of bacterial cell wall assembly proteins | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Marles-Wright, J, Kawai, Y, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Cleverley, R.M, Bui, N.K, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2010-11-11 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Widespread Family of Bacterial Cell Wall Assembly Proteins.
Embo J., 30, 2011
|
|
2XXP
 
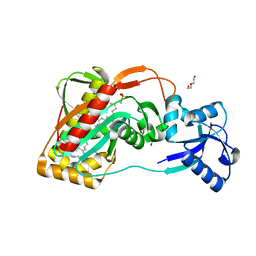 | | A widespread family of bacterial cell wall assembly proteins | | Descriptor: | CPS2A, DI(HYDROXYETHYL)ETHER, MONO-TRANS, ... | | Authors: | Marles-Wright, J, Kawai, Y, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Cleverley, R.M, Bui, N.K, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2010-11-11 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | A Widespread Family of Bacterial Cell Wall Assembly Proteins.
Embo J., 30, 2011
|
|
362D
 
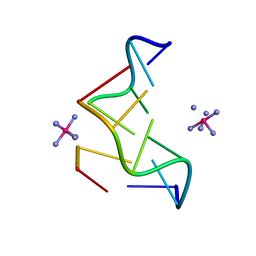 | | THE STRUCTURE OF D(TGCGCA)2 AND A COMPARISON TO OTHER Z-DNA HEXAMERS | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*TP*GP*CP*GP*CP*A)-3') | | Authors: | Harper, N.A, Brannigan, J.A, Buck, M, Lewis, R.J, Moore, M.H, Schneider, B. | | Deposit date: | 1997-08-20 | | Release date: | 1997-11-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of d(TGCGCA)2 and a comparison to other DNA hexamers.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1H4X
 
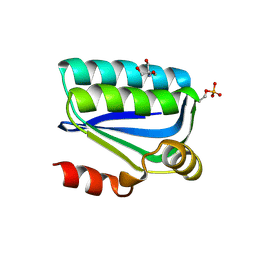 | | Structure of the Bacillus Cell Fate Determinant SpoIIAA in the Phosphorylated Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H4Z
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H4Y
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
4LFT
 
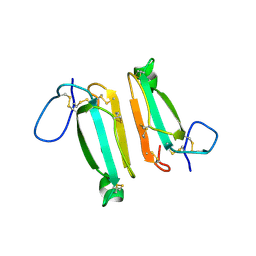 | | Structure of alpha-elapitoxin-Dpp2d isolated from Black Mamba (Dendroaspis polylepis) venom | | Descriptor: | Alpha-elapitoxin-Dpp2a | | Authors: | Wang, C.I.A, Reeks, T, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2013-06-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Isolation and Structural and Pharmacological Characterization of alpha-Elapitoxin-Dpp2d, an Amidated Three Finger Toxin from Black Mamba Venom.
Biochemistry, 53, 2014
|
|
5AN5
 
 | | B. subtilis GpsB C-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB, GLYCEROL | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-09-04 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
2BNJ
 
 | | The xylanase TA from Thermoascus aurantiacus utilizes arabinose decorations of xylan as significant substrate specificity determinants. | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ENDO-1,4-BETA-XYLANASE, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Vardakou, M, Murray, J.W, Flint, J, Christakopoulos, P, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2005-03-25 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Family 10 Thermoascus Aurantiacus Xylanase Utilizes Arabinose Decorations of Xylan as Significant Substrate Specificity Determinants.
J.Mol.Biol., 352, 2005
|
|
2BNL
 
 | |
2C1F
 
 | |
3ZTA
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
3ZQ4
 
 | | Unusual, dual endo- and exo-nuclease activity in the degradosome explained by crystal structure analysis of RNase J1 | | Descriptor: | CALCIUM ION, RIBONUCLEASE J 1, ZINC ION | | Authors: | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A, Harwood, C.R, Lewis, R.J. | | Deposit date: | 2011-06-07 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unusual, Dual Endo- and Exonuclease Activity in the Degradosome Explained by Crystal Structure Analysis of Rnase J1.
Structure, 19, 2011
|
|
3ZTB
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN, IODIDE ION | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
3ZXJ
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXN
 
 | | Moorella thermoacetica RsbS S58E | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN, THIOCYANATE ION | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-08-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
