5GL4
 
 | | Crystal structure of TON_0340 in complex with Mn | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GL2
 
 | | Crystal structure of TON_0340 in complex with Ca | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GKX
 
 | | Crystal structure of TON_0340, apo form | | Descriptor: | PHOSPHATE ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
4FC5
 
 | | Crystal Structure of Ton_0340 | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Lee, S.G, Lee, K.H, An, Y.J, Cha, S.S, Oh, B.H. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8VGB
 
 | |
5XB1
 
 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Ahn, B.J, Jeong, H, Kim, H, Hyun, J, Jung, Y. | | Deposit date: | 2017-03-15 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YI5
 
 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Yoon, H.R, Ahn, B.J, Jeong, H, Hyun, J, Jung, Y, Kim, H. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6E1J
 
 | |
4R6W
 
 | |
4R6X
 
 | |
4S13
 
 | | Ferulic Acid Decarboxylase (FDC1) | | Descriptor: | 4-ethenylphenol, Ferulic acid decarboxylase 1 | | Authors: | Lee, S.G, Bhuiya, M.W, Yu, O, Jez, J.M. | | Deposit date: | 2015-01-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Structure and Mechanism of Ferulic Acid Decarboxylase (FDC1) from Saccharomyces cerevisiae.
Appl.Environ.Microbiol., 81, 2015
|
|
8VGA
 
 | |
5IUU
 
 | |
5IUV
 
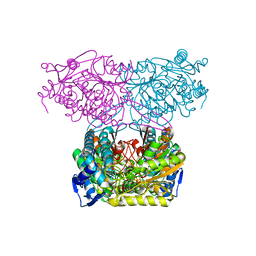 | | Crystal Structure of Indole-3-acetaldehyde Dehydrogenase in complexed with NAD+ | | Descriptor: | Aldehyde dehydrogenase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, S.G, McClerklin, S, Kunkel, B, Jez, J.M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000.
PLoS Pathog., 14, 2018
|
|
5IUW
 
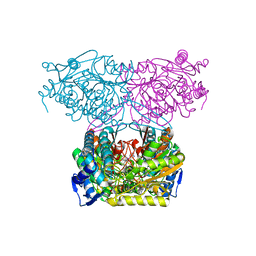 | | Crystal Structure of Indole-3-acetaldehyde Dehydrogenase in complexed with NAD+ and IAA | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Aldehyde dehydrogenase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, S.G, McClerklin, S, Kunkel, B, Jez, J.M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000.
PLoS Pathog., 14, 2018
|
|
5J32
 
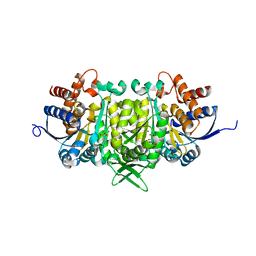 | | Isopropylmalate dehydrogenase in complex with isopropylmalate | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase 2, chloroplastic, ... | | Authors: | Jez, J.M, Lee, S.G. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
5J33
 
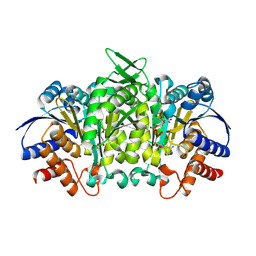 | | Isopropylmalate dehydrogenase in complex with NAD+ | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, MAGNESIUM ION, ... | | Authors: | Jez, J.M, Lee, S.G. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.492 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
6MH4
 
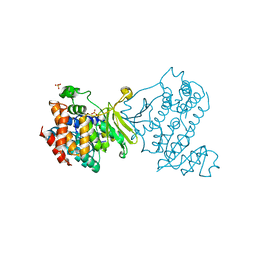 | |
6MH5
 
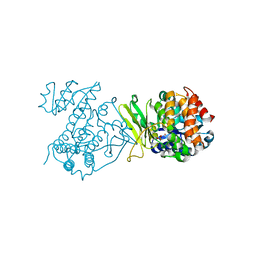 | |
7KRG
 
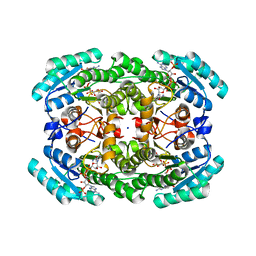 | |
7KQV
 
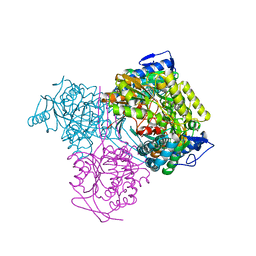 | |
6GSD
 
 | | Plantago Major multifunctional oxidoreductase in complex with progesterone and NADP+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROGESTERONE, Progesterone 5-beta-reductase | | Authors: | Fellows, R, Silva, C, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
4MAF
 
 | | Soybean ATP Sulfurylase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase | | Authors: | Herrmann, J, Ravilious, G.E, McKinney, S.E, Westfall, C.S, Lee, S.G, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and mechanism of soybean ATP sulfurylase and the committed step in plant sulfur assimilation.
J.Biol.Chem., 289, 2014
|
|
5MLM
 
 | | Plantago Major multifunctional oxidoreductase V150M mutant in complex with progesterone and NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, Progesterone 5-beta-reductase | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.563 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
5MLH
 
 | | Plantago Major multifunctional oxidoreductase in complex with 8-oxogeranial and NADP+ | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, CALCIUM ION, GLYCEROL, ... | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-06 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
