1FZW
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). APO ENZYME. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-04 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
2GN9
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-Glc | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2GN4
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADPH and UDP-GlcNAc | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
1RPN
 
 | | Crystal Structure of GDP-D-mannose 4,6-dehydratase in complexes with GDP and NADPH | | Descriptor: | GDP-mannose 4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Webb, N.A, Mulichak, A.M, Lam, J.S, Rocchetta, H.L, Garavito, R.M. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a tetrameric GDP-D-mannose 4,6-dehydratase from a bacterial GDP-D-rhamnose biosynthetic pathway.
Protein Sci., 13, 2004
|
|
2GN8
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2GN6
 
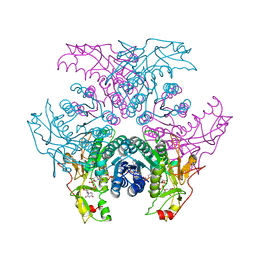 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-GlcNAc | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2GNA
 
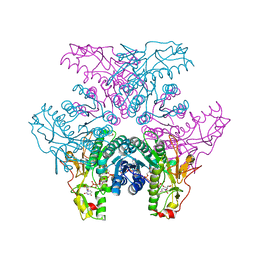 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-Gal | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GALACTOSE-URIDINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
3ME0
 
 | | Structure of the E. coli chaperone PAPD in complex with the pilin domain of the PapGII adhesin | | Descriptor: | Chaperone protein papD, PapG protein | | Authors: | Ford, B.A, Elam, J.S, Dodson, K.W, Pinkner, J.S, Hultgren, S.J. | | Deposit date: | 2010-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the E. Coli Chaperone PapD in Complex with the Pilin Domain of the PapGII Adhesin
To be Published
|
|
2WMP
 
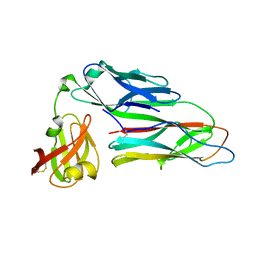 | | Structure of the E. coli chaperone PapD in complex with the pilin domain of the PapGII adhesin | | Descriptor: | CHAPERONE PROTEIN PAPD, PAPG PROTEIN | | Authors: | Ford, B.A, Verger, D, Elam, J.S, Dodson, K.W, Pinkner, J.S, Hultgren, S.J. | | Deposit date: | 2009-07-02 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Papd-Papgii Pilin Complex Reveals an Open and Flexible P5 Pocket.
J.Bacteriol., 194, 2012
|
|
3ODJ
 
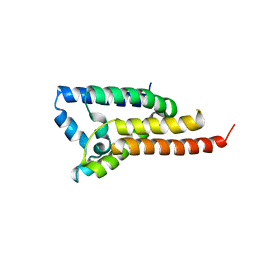 | | Crystal structure of H. influenzae rhomboid GlpG with disordered loop 4, helix 5 and loop 5 | | Descriptor: | Rhomboid protease glpG | | Authors: | Brooks, C.L, Lazareno-Saez, C, Lamoureux, J.S, Mak, M.W, Lemieux, M.J. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Insights into Substrate Gating in H. influenzae Rhomboid.
J.Mol.Biol., 407, 2011
|
|
2IXL
 
 | | RmlC S. suis with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, NICKEL (II) ION | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXC
 
 | | RmlC M. tuberculosis with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE RMLC | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-07 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXJ
 
 | | RmlC P aeruginosa native | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, S,R MESO-TARTARIC ACID | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXI
 
 | | RmlC P aeruginosa with dTDP-xylose | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, S,R MESO-TARTARIC ACID, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXK
 
 | |
2IXH
 
 | | RmlC P aeruginosa with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RmlC, a C3' and C5' carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation.
J. Mol. Biol., 365, 2007
|
|
2PK3
 
 | | Crystal Structure of a GDP-4-keto-6-deoxy-D-mannose reductase | | Descriptor: | GDP-6-deoxy-D-lyxo-4-hexulose reductase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5R)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Webb, N.A, Garavito, R.M. | | Deposit date: | 2007-04-17 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The structural basis for catalytic function of GMD and RMD, two closely related enzymes from the GDP-D-rhamnose biosynthesis pathway.
Febs J., 276, 2009
|
|
3PNT
 
 | | Crystal Structure of the Streptococcus pyogenes NAD+ glycohydrolase SPN in complex with IFS, the Immunity Factor for SPN | | Descriptor: | Immunity factor for SPN, NAD+-glycohydrolase | | Authors: | Smith, C.L, Stine Elam, J, Ellenberger, T, Ghosh, J, Pinkner, J.S, Hultgren, S.J, Caparon, M.G. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Streptococcus pyogenes Immunity to Its NAD(+) Glycohydrolase Toxin.
Structure, 19, 2011
|
|
3QB2
 
 | |
6SQ8
 
 | | Structure of amide bond synthetase McbA from Marinactinospora thermotolerans | | Descriptor: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | Authors: | Rowlinson, B, Petchey, M, Grogan, G. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biocatalytic Synthesis of Moclobemide Using the Amide Bond Synthetase McbA Coupled with an ATP Recycling System.
Acs Catalysis, 10, 2020
|
|
2VSS
 
 | | Wild-type Hydroxycinnamoyl-CoA hydratase lyase in complex with acetyl- CoA and vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
2VSU
 
 | | A ternary complex of Hydroxycinnamoyl-CoA Hydratase-Lyase (HCHL) with acetyl-Coenzyme A and vanillin gives insights into substrate specificity and mechanism. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
6H1B
 
 | | Structure of amide bond synthetase Mcba K483A mutant from Marinactinospora thermotolerans | | Descriptor: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | Authors: | Rowlinson, B, Petchey, M, Cuetos, A, Frese, A, Dannevald, S, Grogan, G. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Broad Aryl Acid Specificity of the Amide Bond Synthetase McbA Suggests Potential for the Biocatalytic Synthesis of Amides.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8PYX
 
 | |
8PPP
 
 | |
