6FH1
 
 | | Protein arginine kinase McsB in the apo state | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, IMIDAZOLE, ... | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
6FH3
 
 | | Protein arginine kinase McsB in the pArg-bound state | | Descriptor: | 1,2-ETHANEDIOL, Protein-arginine kinase, phospho-arginine | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
6FH4
 
 | |
6FH2
 
 | | Protein arginine kinase McsB in the AMP-PN-bound state | | Descriptor: | 1,2-ETHANEDIOL, AMP PHOSPHORAMIDATE, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
3HGS
 
 | | Crystal structure of tomato OPR3 in complex with pHB | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Clausen, T, Breithaupt, C. | | Deposit date: | 2009-05-14 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate specificity of plant 12-oxophytodienoate reductases.
J.Mol.Biol., 392, 2009
|
|
3HGO
 
 | |
3HGR
 
 | | Crystal structure of tomato OPR1 in complex with pHB | | Descriptor: | 12-oxophytodienoate reductase 1, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Clausen, T, Breithaupt, C. | | Deposit date: | 2009-05-14 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of substrate specificity of plant 12-oxophytodienoate reductases.
J.Mol.Biol., 392, 2009
|
|
7AA4
 
 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
7ABR
 
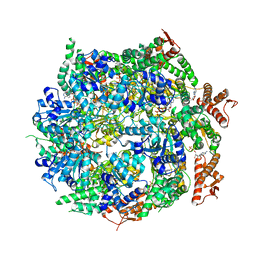 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
2HS6
 
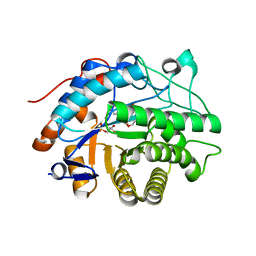 | |
2HS8
 
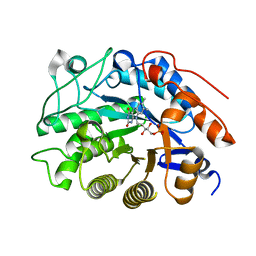 | |
2HSA
 
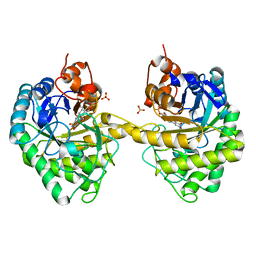 | | Crystal structure of 12-oxophytodienoate reductase 3 (OPR3) from tomato | | Descriptor: | 12-oxophytodienoate reductase 3, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Clausen, T, Huber, R. | | Deposit date: | 2006-07-21 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 12-oxophytodienoate reductase 3 from tomato: Self-inhibition by dimerization.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4RQZ
 
 | |
4RR1
 
 | |
4RR0
 
 | |
2IUN
 
 | | Structure of the C-terminal head domain of the avian adenovirus CELO long fibre (P21 crystal form) | | Descriptor: | AVIAN ADENOVIRUS CELO LONG FIBRE, CALCIUM ION | | Authors: | Guardado-Calvo, P, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J. | | Deposit date: | 2006-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the C-terminal head domain of the fowl adenovirus type 1 long fiber.
J. Gen. Virol., 88, 2007
|
|
2IUM
 
 | |
2INY
 
 | |
