3T7R
 
 | |
3T7T
 
 | |
5AQD
 
 | | Crystal structure of Phormidium Phycoerythrin at pH 8.5 | | Descriptor: | GLYCEROL, PHYCOERYTHRIN ALPHA SUBUNIT, PHYCOERYTHRIN BETA SUBUNIT, ... | | Authors: | Kumar, V, Sharma, M, Sonani, R.R, Gupta, G.D, Madamwar, D. | | Deposit date: | 2015-09-22 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal Structure Analysis of C-Phycoerythrin from Marine Cyanobacterium Phormidium Sp. A09Dm.
Photosynth.Res., 129, 2016
|
|
5A9Y
 
 | | Structure of ppGpp BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9X
 
 | | Structure of GDP bound BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AA0
 
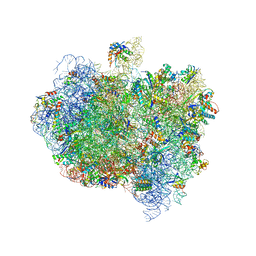 | | Complex of Thermous thermophilus ribosome (A-and P-site tRNA) bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9W
 
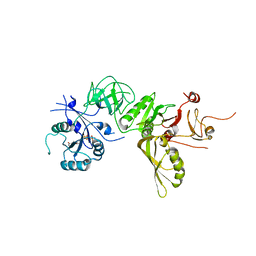 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9V
 
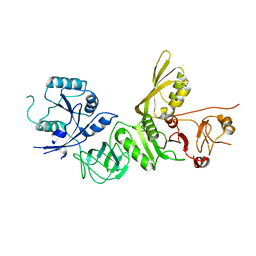 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9Z
 
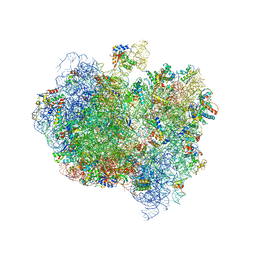 | | Complex of Thermous thermophilus ribosome bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5JCU
 
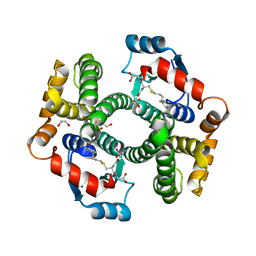 | |
5JCW
 
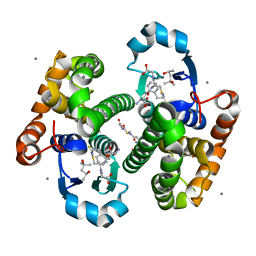 | | Crystal Structure of hGSTP1-1 with Glutathione Adduct of Phenethyl Isothiocyanate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Kumari, V, Ji, X. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Irreversible Inhibition of Glutathione S-Transferase by Phenethyl Isothiocyanate (PEITC), a Dietary Cancer Chemopreventive Phytochemical.
Plos One, 11, 2016
|
|
6ATQ
 
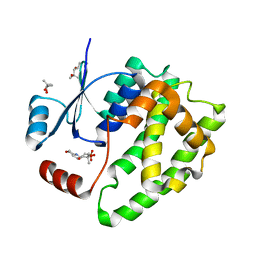 | |
6ATP
 
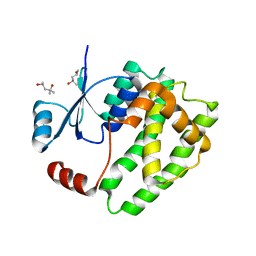 | |
6AP9
 
 | |
6ATO
 
 | |
6ATR
 
 | |
6K5P
 
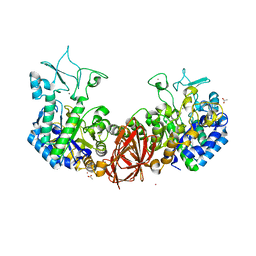 | | Structure of mosquito-larvicidal Binary toxin receptor, Cqm1 | | Descriptor: | ACETATE ION, Binary toxin receptor protein, CADMIUM ION, ... | | Authors: | Kumar, V, Sharma, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Crystal structure of BinAB toxin receptor (Cqm1) protein and molecular dynamics simulations reveal the role of unique Ca(II) ion.
Int.J.Biol.Macromol., 140, 2019
|
|
8SDV
 
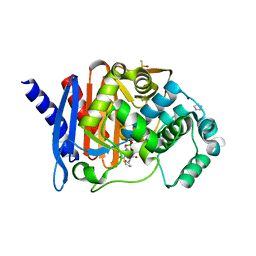 | | Crystal structure of PDC-3 Y221H beta-lactamase in complex with the boronic acid inhibitor S02030 | | Descriptor: | 1-{(2R)-2-(dihydroxyboranyl)-2-[(thiophen-2-ylacetyl)amino]ethyl}-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8SDR
 
 | |
8SDN
 
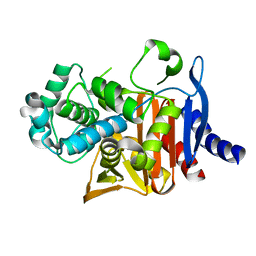 | | Crystal structure of PDC-3 Y221H beta-lactamase | | Descriptor: | Beta-lactamase, ISOPROPYL ALCOHOL | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8SDL
 
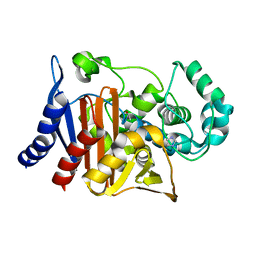 | | Crystal structure of PDC-3 beta-lactamase | | Descriptor: | Beta-lactamase, IMIDAZOLE, ISOPROPYL ALCOHOL | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8SDT
 
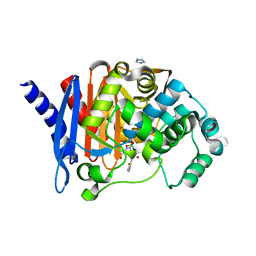 | |
8SDS
 
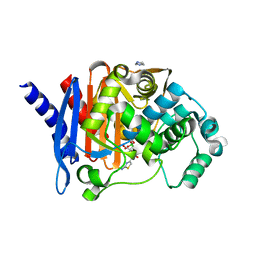 | |
1KSI
 
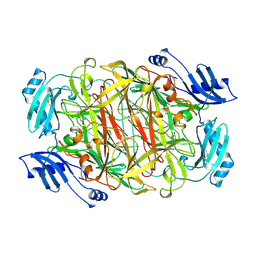 | | CRYSTAL STRUCTURE OF A EUKARYOTIC (PEA SEEDLING) COPPER-CONTAINING AMINE OXIDASE AT 2.2A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Wilce, M.C.J, Kumar, V, Freeman, H.C, Guss, J.M. | | Deposit date: | 1996-07-20 | | Release date: | 1997-12-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 A resolution.
Structure, 4, 1996
|
|
6VJE
 
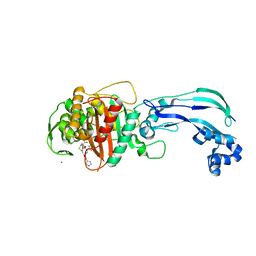 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
