5GT3
 
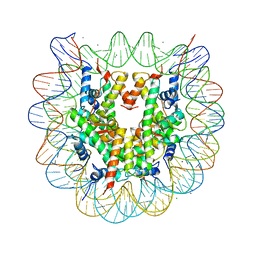 | | Crystal structure of nucleosome particle in the presence of human testis-specific histone variant, hTh2b | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-D, ... | | Authors: | Kumarevel, T, Sivaraman, P. | | Deposit date: | 2016-08-18 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural analyses of the nucleosome complexes with human testis-specific histone variants, hTh2a and hTh2b
Biophys. Chem., 221, 2017
|
|
3AOI
 
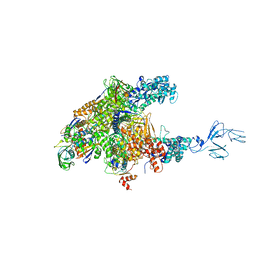 | | RNA polymerase-Gfh1 complex (Crystal type 2) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*A*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
3AOH
 
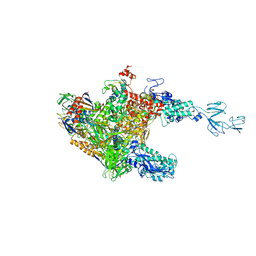 | | RNA polymerase-Gfh1 complex (Crystal type 1) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*AP*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-28 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
2EF7
 
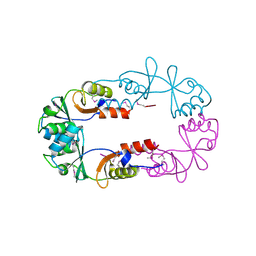 | | Crystal structure of ST2348, a hypothetical protein with CBS domains from Sulfolobus tokodaii strain7 | | Descriptor: | Hypothetical protein ST2348 | | Authors: | Agari, Y, Karthe, P, Kumarevel, T, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-21 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ST2348, a CBS domain protein, from hyperthermophilic archaeon Sulfolobus tokodaii
Biochem.Biophys.Res.Commun., 375, 2008
|
|
3BOY
 
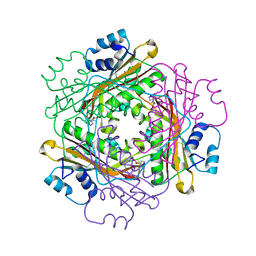 | | Crystal structure of the HutP antitermination complex bound to the HUT mRNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*U)-3', HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T.S, Balasundaresan, D, Jeyakanthan, J, Shinkai, A, Yokoyama, S, Kumar, P.K.R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of HutP complexed with the 55-mer RNA
To be Published
|
|
1WMQ
 
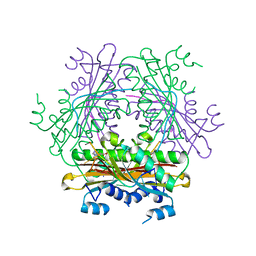 | | Structure of the HutP antitermination complex bound to a single stranded region of hut mRNA | | Descriptor: | 5'-R(P*UP*UP*UP*AP*GP*UP*U)-3', HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-07-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand.
Nature, 434, 2005
|
|
1WPV
 
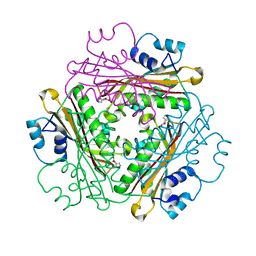 | | Crystal Structure of Activated Binary complex of HutP, an RNA binding anti-termination protein | | Descriptor: | HISTIDINE, Hut operon positive regulatory protein, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand.
Nature, 434, 2005
|
|
1WPS
 
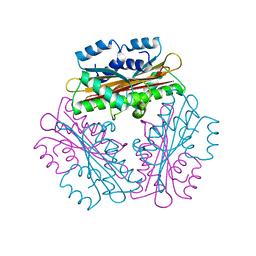 | |
2EB7
 
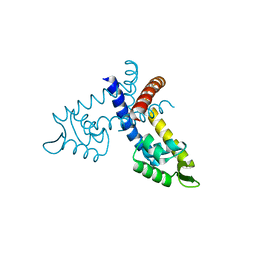 | | Crystal structure of the hypothetical regulator from Sulfolobus tokodaii 7 | | Descriptor: | 146aa long hypothetical transcriptional regulator | | Authors: | Kumarevel, T.S, Nishio, M, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the MarR family regulatory protein, ST1710, from Sulfolobus tokodaii strain 7
J.Struct.Biol., 161, 2008
|
|
2E85
 
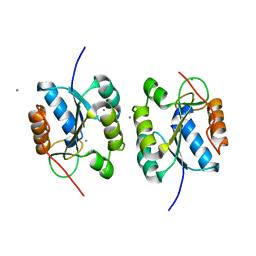 | | Crystal Structure of the Hydrogenase 3 Maturation protease | | Descriptor: | CALCIUM ION, Hydrogenase 3 maturation protease | | Authors: | Tanaka, T, Kumarevel, T.S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-18 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of hydrogenase maturating endopeptidase HycI from Escherichia coli
Biochem.Biophys.Res.Commun., 389, 2009
|
|
7LLB
 
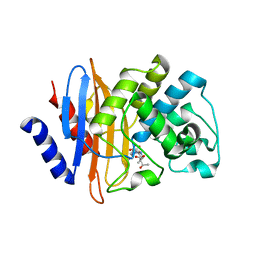 | | Crystal structure of KPC-2 S70G/T215P mutant with hydrolyzed meropenem | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LLH
 
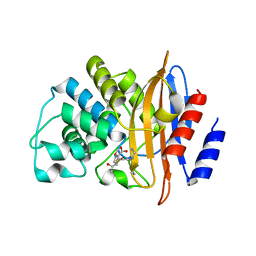 | | KPC-2 F72Y mutant with acylated imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LJK
 
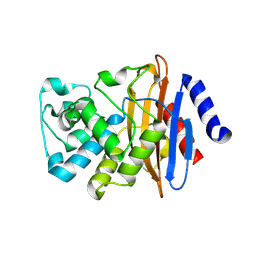 | | Crystal structure of the deacylation deficient KPC-2 F72Y mutant | | Descriptor: | Beta-lactamase | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
6B1J
 
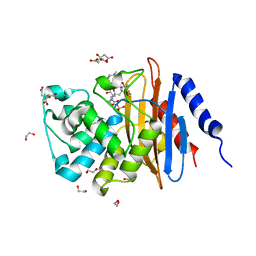 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 5107 by soaking | | Descriptor: | (2S,5R)-1-formyl-N'-[(3R)-piperidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-18 | | Release date: | 2018-08-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
6B1F
 
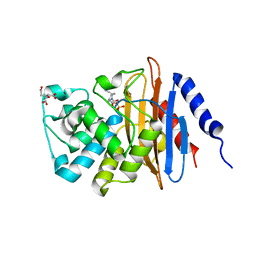 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 4234 by soaking | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-18 | | Release date: | 2018-08-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
6B1H
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 4234 by co-crystallization | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, ACETATE ION, CHLORIDE ION, ... | | Authors: | van den Akker, F, Nhuyen, N.Q. | | Deposit date: | 2017-09-18 | | Release date: | 2018-08-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
6B1W
 
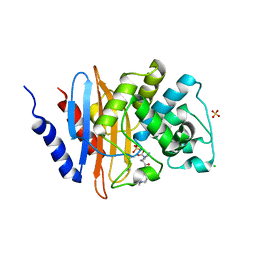 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 5107 by co-crystallization | | Descriptor: | (2S,5R)-1-formyl-N'-[(3R)-piperidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, CHLORIDE ION, Carbapenem-hydrolyzing beta-lactamase KPC, ... | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
6B22
 
 | | Crystal structure OXA-24 beta-lactamase complexed with WCK 4234 by co-crystallization | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, Beta-lactamase, CHLORIDE ION | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
6B1Y
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 5153 by co-crystallization | | Descriptor: | (2S,5Z)-1-formyl-5-imino-N'-[(3R)-1-(sulfooxy)pyrrolidine-3-carbonyl]piperidine-2-carbohydrazide, CHLORIDE ION, Carbapenem-hydrolyzing beta-lactamase KPC, ... | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
6B1X
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 5153 by soaking | | Descriptor: | (2S,5R)-1-formyl-N'-[(3R)-pyrrolidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
3UV1
 
 | | Crystal structure a major allergen from dust mite | | Descriptor: | Der f 7 allergen | | Authors: | Tan, K.W, Kumar, T, Chew, F.T, Mok, Y.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Der f 7, a Dust Mite Allergen from Dermatophagoides farinae.
Plos One, 7, 2012
|
|
1Y6L
 
 | | Human ubiquitin conjugating enzyme E2E2 | | Descriptor: | Ubiquitin-conjugating enzyme E2E2 | | Authors: | Walker, J.R, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Bochkarev, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
1YH2
 
 | | Ubiquitin-Conjugating Enzyme HSPC150 | | Descriptor: | HSPC150 protein similar to ubiquitin-conjugating enzyme | | Authors: | Walker, J.R, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
3X1U
 
 | |
3X1V
 
 | |
