3C5O
 
 | | Crystal structure of the conserved protein of unknown function RPA1785 from Rhodopseudomonas palustris | | Descriptor: | GLYCEROL, UPF0311 protein RPA1785 | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-01 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Conserved Protein of Unknown Function RPA1785 from Rhodopseudomonas palustris.
To be Published
|
|
1XPP
 
 | | Crystal Structure of TA1416,DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum | | Descriptor: | ACETIC ACID, DNA-directed RNA polymerase subunit L, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of TA1416, DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum
To be Published
|
|
3EFB
 
 | | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri | | Descriptor: | ACETIC ACID, Probable sor-operon regulator | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri
To be Published
|
|
3EC7
 
 | | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETIC ACID, ... | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2
To be Published
|
|
1SED
 
 | | Crystal Structure of Protein of Unknown Function YhaL from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Hypothetical protein yhaI, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Hypothetical Protein YhaI, APC1180 from Bacillus subtilis
To be Published
|
|
5TF0
 
 | | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 3 N-terminal domain protein, MAGNESIUM ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.2021 Å) | | Cite: | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis
To Be Published, 2016
|
|
5TK4
 
 | | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein from Bacillus anthracis | | Descriptor: | Cytochrome B | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein from
Bacillus anthracis
To Be Published
|
|
5TF3
 
 | | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, Putative membrane protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Anderson, W.F, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis
To Be Published
|
|
5TK2
 
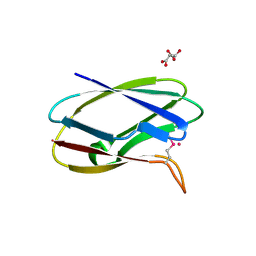 | | Crystal Structure of Uncharacterized Cupredoxin-like domain protein from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Cytochrome B, ... | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Uncharacterized Cupredoxin-like domain protein from
Bacillus anthracis
To Be Published
|
|
1TE2
 
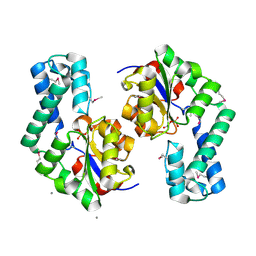 | | Putative Phosphatase Ynic from Escherichia coli K12 | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 2-deoxyglucose-6-P phosphatase, CALCIUM ION | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Putative Phosphatase Ynic from Escherichia coli K12
To be Published
|
|
4Q63
 
 | | Crystal Structure of Legionella Uncharacterized Protein Lpg0364 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-21 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Crystal Structure of Legionella Uncharacterized Protein Lpg0364
To be Published
|
|
9C3D
 
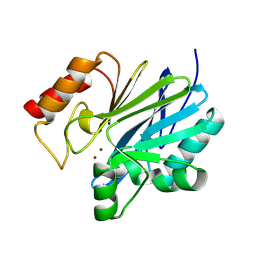 | | Crystal structure of metallo-beta-lactamase superfamily protein CcrA-like_MBL-B1 from Dyadobacter fermentans | | Descriptor: | Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kim, Y, Maltseva, N, Welk, L, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-31 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of metallo-beta-lactamase superfamily protein CcrA-like_MBL-B1 from Dyadobacter fermentans
To Be Published
|
|
9BZB
 
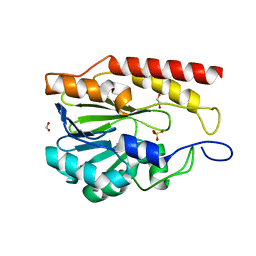 | | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SULFATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2
To Be Published
|
|
9DCG
 
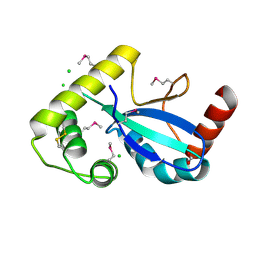 | | Crystal Structure of the Thiol:Disulfide Interchange Protein DsbC from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the Thiol:Disulfide Interchange Protein DsbC from Vibrio cholerae
To Be Published
|
|
9DDY
 
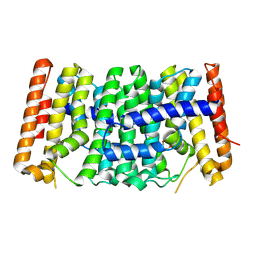 | | The Crystal Structure of Geranyltranstransferase from Streptococcus pneumoniae TIGR4 | | Descriptor: | GLYCEROL, Geranyltranstransferase | | Authors: | Kim, Y, Tan, A, Endres, M, Tan, K, Joachimik, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-28 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The Crystal Structure of Geranyltranstransferase from Streptococcus pneumoniae TIGR4
To Be Published
|
|
9GTE
 
 | | Crystal structure of TRIM21 PRY-SPRY domain bound to Suramin | | Descriptor: | 1,2-ETHANEDIOL, 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Kim, Y, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-17 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of TRIM21 PRY-SPRY domain bound to Suramin
To Be Published
|
|
9D5W
 
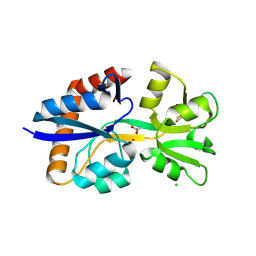 | | Crystal Structure of the Substrate Binding Domain Protein of the ABC Transporter PBP2_YxeM from Vibrio cholerae | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding portion, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Grimshaw, S, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-14 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Substrate Binding Domain Protein of the ABC Transporter PBP2_YxeM from Vibrio cholerae
To Be Published
|
|
2G7G
 
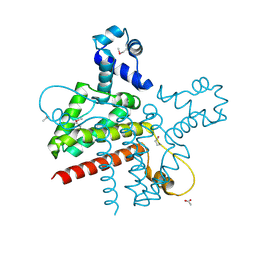 | | The Crystal Structure of the Putative Transcriptional Regulator Rha04620 from Rhodococcus sp. RHA1 | | Descriptor: | ACETIC ACID, Rha04620, Putative Transcriptional Regulator | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Crystal Structure of the Putative Transcriptional Regulator Rha04620 from Rhodococcus sp. RHA1
To be Published
|
|
1EUJ
 
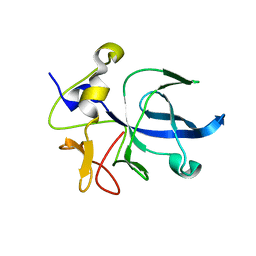 | | A NOVEL ANTI-TUMOR CYTOKINE CONTAINS A RNA-BINDING MOTIF PRESENT IN AMINOACYL-TRNA SYNTHETASES | | Descriptor: | ENDOTHELIAL MONOCYTE ACTIVATING POLYPEPTIDE 2 | | Authors: | Kim, Y, Shin, J, Li, R, Cheong, C, Kim, S. | | Deposit date: | 2000-04-17 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel anti-tumor cytokine contains an RNA binding motif present in aminoacyl-tRNA synthetases.
J.Biol.Chem., 275, 2000
|
|
6WXC
 
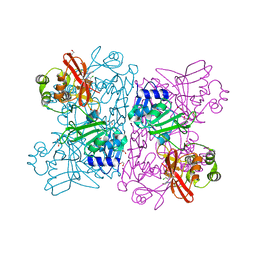 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WLC
 
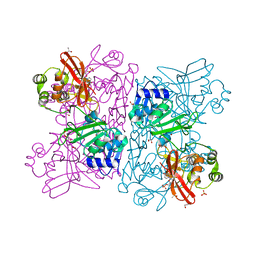 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6X1B
 
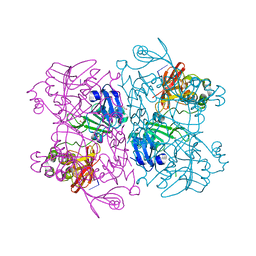 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-18 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
1FM2
 
 | | THE 2 ANGSTROM CRYSTAL STRUCTURE OF CEPHALOSPORIN ACYLASE | | Descriptor: | GLUTARYL 7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Kim, Y, Yoon, K.H, Khang, Y, Turley, S, Hol, W.G.J. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of cephalosporin acylase.
Structure Fold.Des., 8, 2000
|
|
5L07
 
 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Quorum-sensing transcriptional activator, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5KWS
 
 | | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-19 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose
To Be Published
|
|
