6K5J
 
 | | Structure of a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GH3 beta-N-acetylglucosaminidase, GLYCEROL | | Authors: | Itoh, T, Araki, T, Nishiyama, T, Hibi, T, Kimoto, H. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural and functional characterization of a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7.
J.Biochem., 166, 2019
|
|
2D8L
 
 | | Crystal Structure of Unsaturated Rhamnogalacturonyl Hydrolase in complex with dGlcA-GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2005-12-06 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide
J.Mol.Biol., 360, 2006
|
|
2FUZ
 
 | | UGL hexagonal crystal structure without glycine and DTT molecules | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
6K0M
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11 | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, GLYCEROL, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0U
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
2FV0
 
 | | UGL_D88N/dGlcA-Glc-Rha-Glc | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-beta-D-glucopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-beta-D-glucopyranose, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2FV1
 
 | | UGL_D88N/dGlcA-GlcNAc | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
6K0Q
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0S
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0V
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0N
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
2GH4
 
 | | YteR/D143N/dGalA-Rha | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-03-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate
Biochem.Biophys.Res.Commun., 347, 2006
|
|
6KFN
 
 | | Crystal structure of alginate lyase from Paenibacillus sp. str. FPU-7 | | Descriptor: | IMIDAZOLE, SODIUM ION, alginate lyase | | Authors: | Itoh, T, Nakagawa, E, Yoda, M, Nakaichi, A, Hibi, T, Kimoto, H. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural and biochemical characterisation of a novel alginate lyase from Paenibacillus sp. str. FPU-7.
Sci Rep, 9, 2019
|
|
6K0P
 
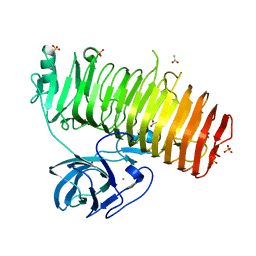 | | Catalytic domain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
2D5J
 
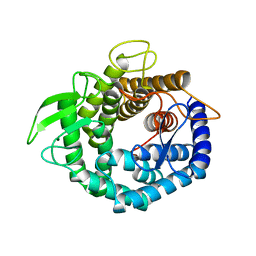 | |
2ZBL
 
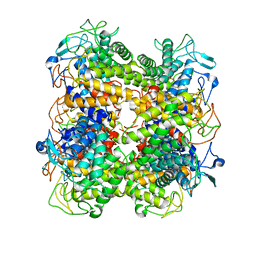 | | Functional annotation of Salmonella enterica yihS-encoded protein | | Descriptor: | Putative isomerase, beta-D-mannopyranose | | Authors: | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
3A09
 
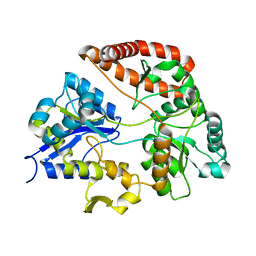 | |
3AX3
 
 | |
3AX2
 
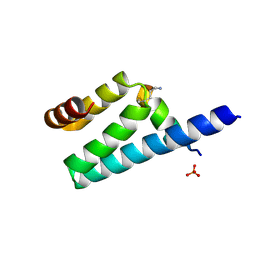 | | Crystal structure of rat TOM20-ALDH presequence complex: a disulfide-tethered complex with a non-optimized, long linker | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
3AX5
 
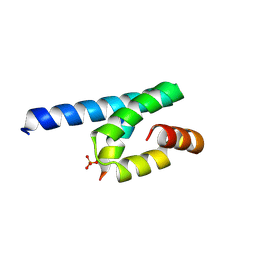 | | Crystal structure of rat TOM20-ALDH presequence complex: A complex (form1) between Tom20 and a disulfide-bridged presequence peptide containing D-Cys and L-Cys at the i and i+3 positions. | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
5Y52
 
 | |
5Y2P
 
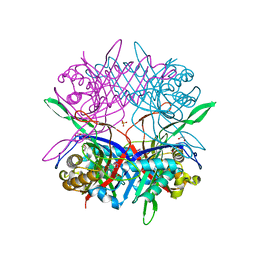 | |
2MWI
 
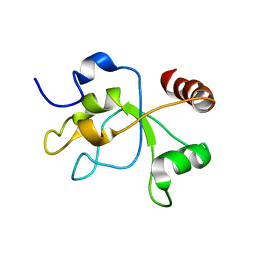 | | The structure of the carboxy-terminal domain of DNTTIP1 | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1 | | Authors: | Schwabe, J.W.R, Muskett, F.W, Itoh, T. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting.
Nucleic Acids Res., 43, 2015
|
|
2D5F
 
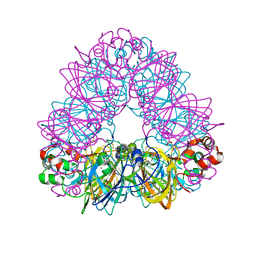 | | Crystal Structure of Recombinant Soybean Proglycinin A3B4 subunit, its Comparison with Mature Glycinin A3B4 subunit, Responsible for Hexamer Assembly | | Descriptor: | CARBONATE ION, MAGNESIUM ION, glycinin A3B4 subunit | | Authors: | Itoh, T, Adachi, M, Masuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2005-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
2D5H
 
 | | Crystal Structure of Recombinant Soybean Proglycinin A3B4 subunit, its Comparison with Mature Glycinin A3B4 subunit, Responsible for Hexamer Assembly | | Descriptor: | CARBONATE ION, MAGNESIUM ION, glycinin A3B4 subunit | | Authors: | Itoh, T, Adachi, M, Masuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2005-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
