7L59
 
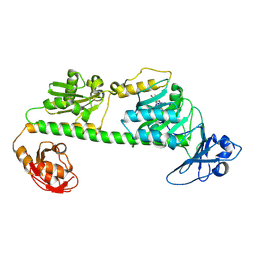 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP-G454E variant from Xanthomonas campestris in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
7KVC
 
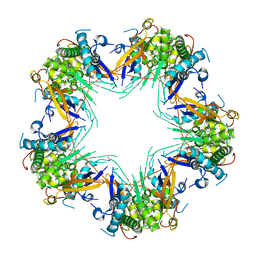 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7KVD
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
5M7P
 
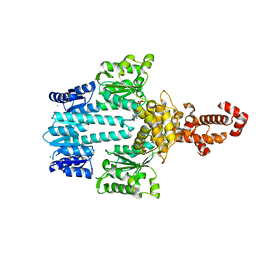 | | Crystal structure of NtrX from Brucella abortus in complex with ADP processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
5M7O
 
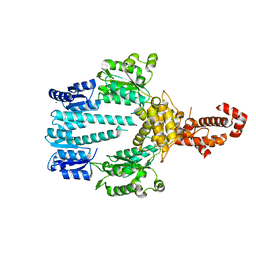 | | Crystal structure of NtrX from Brucella abortus processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
2O6H
 
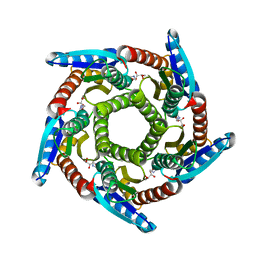 | | Lumazine synthase RibH1 from Brucella melitensis (Gene BMEI1187, Swiss-Prot entry Q8YGH2) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-12-07 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
2OBX
 
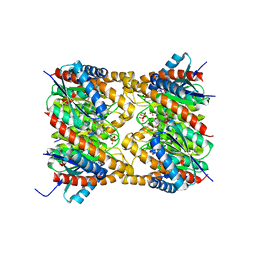 | | Lumazine synthase RibH2 from Mesorhizobium loti (Gene mll7281, Swiss-Prot entry Q986N2) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, PHOSPHATE ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-12-20 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and kinetic properties of lumazine synthase isoenzymes in the order rhizobiales
J.Mol.Biol., 373, 2007
|
|
5M7N
 
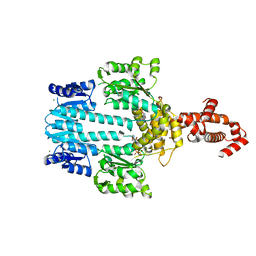 | | Crystal structure of NtrX from Brucella abortus in complex with ATP processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
4FXU
 
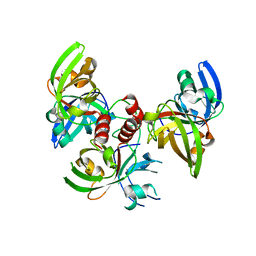 | | Crystallographic structure of trimeric riboflavin synthase from Brucella abortus | | Descriptor: | Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-07-03 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4G6I
 
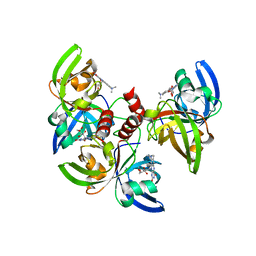 | | Crystallographic structure of trimeric riboflavin synthase from Brucella abortus in complex with roseoflavin | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-07-19 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4G97
 
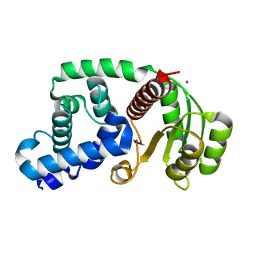 | | Crystal structure of the response regulator PhyR from Brucella abortus | | Descriptor: | CALCIUM ION, Response regulator receiver | | Authors: | Klinke, S, Sycz, G, Rinaldi, J.J, Paris, G, Goldbaum, F.A. | | Deposit date: | 2012-07-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the response regulator PhyR from Brucella abortus
To be Published
|
|
4E0F
 
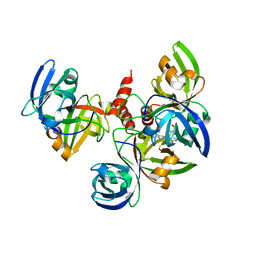 | | Crystallographic structure of trimeric Riboflavin Synthase from Brucella abortus in complex with riboflavin | | Descriptor: | RIBOFLAVIN, Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-03-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4GQN
 
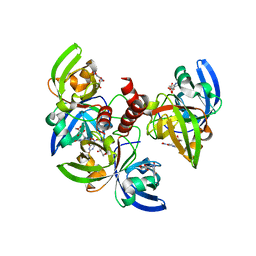 | | Crystallographic structure of trimeric Riboflavin Synthase from Brucella abortus in complex with 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-08-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6UCT
 
 | | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Klinke, S, Monti, D, Sycz, G, Cerutti, M.L, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2019-09-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant)
To Be Published
|
|
6V95
 
 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-tartaramidoyl derivative (diNGT) | | Descriptor: | (2R,3R)-N-[(1-{(3S,3aR,6S,6aR)-6-[4-({[(2R,3R)-2,3-dihydroxy-4-oxo-4-{[(2R,3R,4R,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]amino}butanoyl]amino}methyl)-1H-1,2,3-triazol-1-yl]hexahydrofuro[3,2-b]furan-3-yl}-1H-1,2,3-triazol-4-yl)methyl]-2,3-dihydroxy-N'-[(2R,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]butanediamide (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-13 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VGF
 
 | | Peanut lectin complexed with divalent S-beta-D-thiogalactopyranosyl beta-D-glucopyranoside derivative (diSTGD) | | Descriptor: | (2S,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6R)-3,4,5-trihydroxy-6-{[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-({4-[({[(2S,3S,4S,5R,6S)-3,4,5-trihydroxy-6-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-2-yl]methyl}sulfanyl)methyl]-1H-1,2,3-triazol-1-yl}methyl)tetrahydro-2H-pyran-2-yl]oxy}tetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VAW
 
 | | Peanut lectin complexed with N-beta-D-galactopyranosyl-L-succinamoyl derivative (NGS) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC4
 
 | | Peanut lectin complexed with S-beta-D-Thiogalactopyranosyl beta-D-glucopyranoside derivative (STGD) | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6S)-3,4,5-trihydroxy-6-methoxytetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VAV
 
 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-succinamoyl derivative (diNGS) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC3
 
 | | Peanut lectin complexed with S-beta-D-thiogalactopyranosyl 6-deoxy-6-S-propynyl-beta-D-glucopyranoside (STG) | | Descriptor: | 6-S-(prop-2-yn-1-yl)-6-thio-beta-D-glucopyranosyl 1-thio-beta-D-galactopyranoside, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1EMT
 
 | | FAB ANTIBODY FRAGMENT OF AN C60 ANTIFULLERENE ANTIBODY | | Descriptor: | IGG ANTIBODY (HEAVY CHAIN), IGG ANTIBODY (LIGHT CHAIN) | | Authors: | Braden, B.C, Goldbaum, F.A, Chen, B.-X, Erlanger, B.F, Kirschner, A.N, Wilson, S.R. | | Deposit date: | 2000-03-17 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystal structure of an anti-Buckminsterfullerene antibody fab fragment: biomolecular recognition of C(60).
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1P2C
 
 | |
1QLR
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF A HUMAN MONOCLONAL IgM COLD AGGLUTININ | | Descriptor: | IGM FAB REGION IV-J(H4)-C (KAU COLD AGGLUTININ), IGM KAPPA CHAIN V-III (KAU COLD AGGLUTININ), alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carvalho, J.G, Cauerhff, A, Goldbaum, F, Leoni, J, Polikarpov, I. | | Deposit date: | 1999-09-11 | | Release date: | 2000-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Three-dimensional structure of the Fab from a human IgM cold agglutinin.
J Immunol., 165, 2000
|
|
1KIP
 
 | |
1KIQ
 
 | |
