4XV6
 
 | | CcP gateless cavity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XVA
 
 | | Crystal structure of wild type cytochrome c peroxidase | | Descriptor: | BENZIMIDAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XV4
 
 | | CcP gateless cavity | | Descriptor: | 2-AMINO-5-METHYLTHIAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
7MFT
 
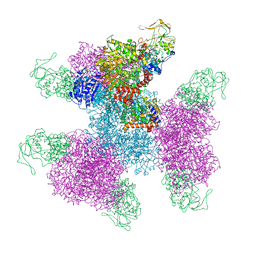 | | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex (GudB6-GltA6-GltB6) | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Jayaraman, V, Lee, D.J, Elad, N, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2021-04-11 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Nat.Chem.Biol., 18, 2022
|
|
7MFM
 
 | | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Jayaraman, V, Lee, D.J, Elad, N, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2021-04-10 | | Release date: | 2022-01-05 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Nat.Chem.Biol., 18, 2022
|
|
4NVA
 
 | |
4NVL
 
 | |
4NVH
 
 | |
4OQ7
 
 | |
4OQI
 
 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying R164S/G238S mutations | | Descriptor: | CALCIUM ION, SULFATE ION, TEM-94 ES-beta-lactamase | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-09 | | Release date: | 2015-05-20 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
4OP8
 
 | | Room temperature crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying G238S mutation | | Descriptor: | CALCIUM ION, SULFATE ION, TEM-94 ES-beta-lactamase, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-05 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
4OPQ
 
 | | Room temperature crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying R164S/G238S mutations | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
4OPZ
 
 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying G238S mutation in complex with boron-based inhibitor EC25 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2R)-2-{[(2R)-2-amino-2-phenylacetyl]amino}-2-(dihydroxyboranyl)ethyl]benzoic acid, ACETATE ION, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-07 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
5KV7
 
 | | Human cyclophilin A at 278K, Data set 9 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KW7
 
 | | T. danielli thaumatin at 278K, Data set 4 | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S. | | Deposit date: | 2016-07-15 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXK
 
 | | Hen Egg White Lysozyme at 100K, Data set 1 | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXP
 
 | | Hen Egg White Lysozyme at 278K, Data set 2 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXY
 
 | | Hen Egg White Lysozyme at 278K, Data set 8 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KUL
 
 | | Human cyclophilin A at 100K, Data set 1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KUU
 
 | | Human cyclophilin A at 100K, Data set 7 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KV2
 
 | | Human cyclophilin A at 278K, Data set 4 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KUS
 
 | | Human cyclophilin A at 100K, Data set 6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KV1
 
 | | Human cyclophilin A at 278K, Data set 3 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXT
 
 | | Hen Egg White Lysozyme at 278K, Data set 5 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KVW
 
 | | T. danielli thaumatin at 100K, Data set 1 | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
