2F9A
 
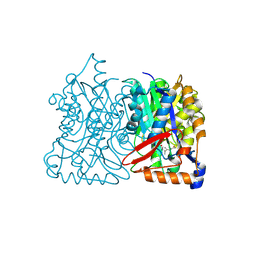 | | HMG-CoA synthase from Brassica juncea in complex with F-244 | | Descriptor: | (7R,12R,13R)-13-formyl-12,14-dihydroxy-3,5,7-trimethyltetradeca-2,4-dienoic acid, 3-Hydroxy-3-methylglutaryl coenzyme A synthase 1 | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-05 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FA3
 
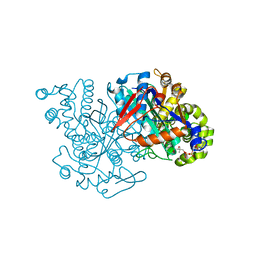 | | HMG-CoA synthase from Brassica juncea in complex with acetyl-CoA and acetyl-cys117. | | Descriptor: | ACETYL COENZYME *A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2F82
 
 | |
2FA0
 
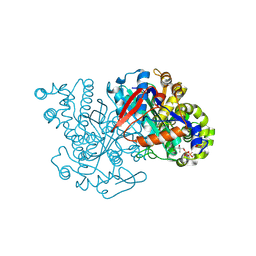 | | HMG-CoA synthase from Brassica juncea in complex with HMG-CoA and covalently bound to HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2CDQ
 
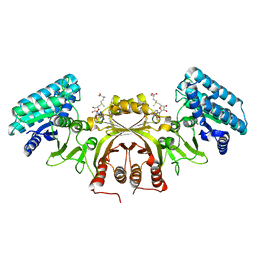 | | Crystal structure of Arabidopsis thaliana aspartate kinase complexed with lysine and S- adenosylmethionine | | Descriptor: | ASPARTOKINASE, D(-)-TARTARIC ACID, LYSINE, ... | | Authors: | Mas-Droux, C, Curien, G, Robert-Genthon, M, Laurencin, M, Ferrer, J.L, Dumas, R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Novel Organization of Act Domains in Allosteric Enzymes Revealed by the Crystal Structure of Arabidopsis Aspartate Kinase
Plant Cell, 18, 2006
|
|
1DDI
 
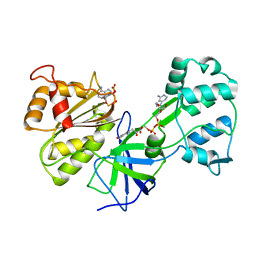 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFITE REDUCTASE [NADPH] FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1DDG
 
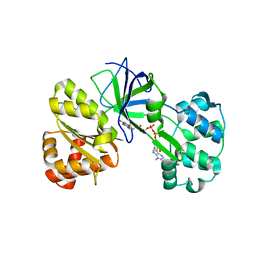 | | CRYSTAL STRUCTURE OF SIR-FP60 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, SULFITE REDUCTASE (NADPH) FLAVOPROTEIN ALPHA-COMPONENT | | Authors: | Gruez, A, Pignol, D, Zeghouf, M, Coves, J, Fontecave, M, Ferrer, J.L, Fontecilla-Camps, J.C. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Four crystal structures of the 60 kDa flavoprotein monomer of the sulfite reductase indicate a disordered flavodoxin-like module.
J.Mol.Biol., 299, 2000
|
|
1Y6I
 
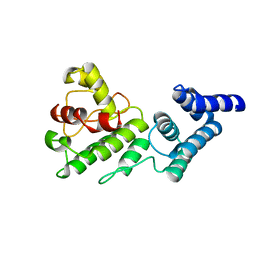 | | Synechocystis GUN4 | | Descriptor: | Mg-chelatase cofactor GUN4 | | Authors: | Verdecia, M.A, Larkin, R.M, Ferrer, J.L, Riek, R, Chory, J, Noel, J.P. | | Deposit date: | 2004-12-06 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding
Plos Biol., 3, 2005
|
|
2BKW
 
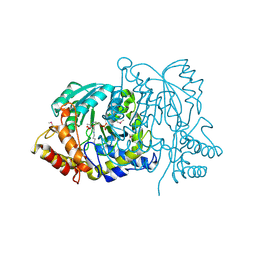 | | Yeast alanine:glyoxylate aminotransferase YFL030w | | Descriptor: | ALANINE-GLYOXYLATE AMINOTRANSFERASE 1, GLYOXYLIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meyer, P, Liger, D, Leulliot, N, Quevillon-Cheruel, S, Zhou, C.Z, Borel, F, Ferrer, J.L, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2005-02-21 | | Release date: | 2005-11-02 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure and Confirmation of the Alanine:Glyoxylate Aminotransferase Activity of the Yfl030W Yeast Protein
Biochimie, 87, 2005
|
|
3ZI7
 
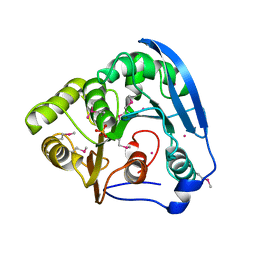 | |
1CGZ
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH RESVERATROL | | Descriptor: | PROTEIN (CHALCONE SYNTHASE), RESVERATROL | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-26 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CGK
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH NARINGENIN | | Descriptor: | NARINGENIN, PROTEIN (CHALCONE SYNTHASE) | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-25 | | Release date: | 1999-09-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CML
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH MALONYL-COA | | Descriptor: | MALONYL-COENZYME A, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), PROTEIN (CHALCONE SYNTHASE), ... | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1CHW
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH HEXANOYL-COA | | Descriptor: | HEXANOYL-COENZYME A, PROTEIN (CHALCONE SYNTHASE) | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1BQ6
 
 | | CHALCONE SYNTHASE FROM ALFALFA WITH COENZYME A | | Descriptor: | CHALCONE SYNTHASE, COENZYME A, SULFATE ION | | Authors: | Ferrer, J.-L, Bowman, M.E, Jez, J, Dixon, R, Noel, J.P. | | Deposit date: | 1998-08-21 | | Release date: | 1999-08-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
3ZI6
 
 | |
4QVJ
 
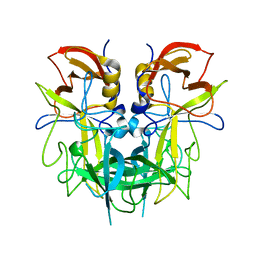 | |
4QV2
 
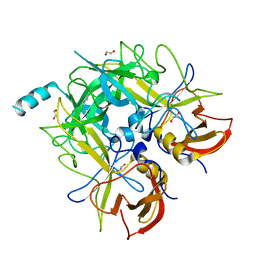 | |
2WGV
 
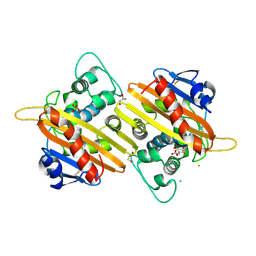 | | Crystal structure of the OXA-10 V117T mutant at pH 6.5 inhibited by a chloride ion | | Descriptor: | BETA-LACTAMASE OXA-10, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post- Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
5YRS
 
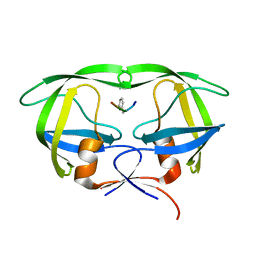 | | X-ray Snapshot of HIV-1 Protease in Action: Observation of Tetrahedral Intermediate and Its SIHB with Catalytic Aspartate | | Descriptor: | PROTEASE, RT-RH oligopeprtide | | Authors: | Das, A, Mahale, S, Prashar, V, Bihani, S, Ferrer, J.-L, Hosur, M.V. | | Deposit date: | 2017-11-10 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray snapshot of HIV-1 protease in action: observation of tetrahedral intermediate and short ionic hydrogen bond SIHB with catalytic aspartate.
J. Am. Chem. Soc., 132, 2010
|
|
3BWN
 
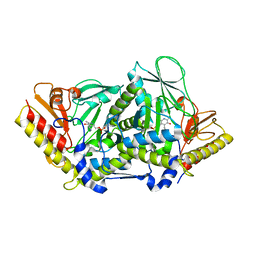 | | L-tryptophan aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-tryptophan aminotransferase, PHENYLALANINE, ... | | Authors: | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | Deposit date: | 2008-01-10 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants
Cell(Cambridge,Mass.), 133, 2008
|
|
1KYW
 
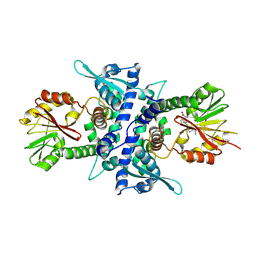 | | Crystal Structure Analysis of Caffeic Acid/5-hydroxyferulic acid 3/5-O-methyltransferase in complex with 5-hydroxyconiferaldehyde | | Descriptor: | 5-(3,3-DIHYDROXYPROPENY)-3-METHOXY-BENZENE-1,2-DIOL, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
1KYZ
 
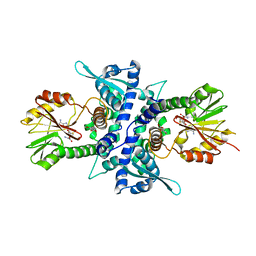 | | Crystal Structure Analysis of Caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase Ferulic Acid Complex | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
3QYZ
 
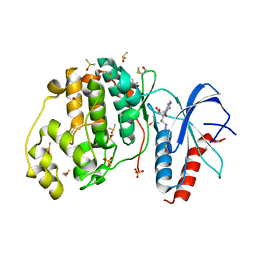 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 5'-azido-8-bromo-5'-deoxyadenosine, BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3QYW
 
 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 6-(3-bromophenyl)-7H-purin-2-amine, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Pochet, S, Hoh, F, Pirochi, M, Guichou, J.-F, Ferrer, J.-L, Labesse, G. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
