2NQI
 
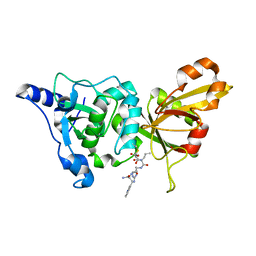 | | Calpain 1 proteolytic core inactivated by WR13(R,R), an epoxysuccinyl-type inhibitor. | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N~2~-[(2S)-2-{[(2R)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]AMINO}PENT-4-ENOYL]-L-ARGINYL-L-TRYPTOPHANAMIDE | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
2NQG
 
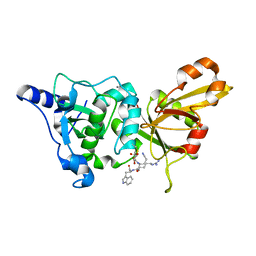 | | Calpain 1 proteolytic core inactivated by WR18(S,S), an epoxysuccinyl-type inhibitor. | | Descriptor: | 5-AZANYLIDYNE-N-[(2S)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]-L-NORVALYL-L-ARGINYL-L-TRYPTOPHANAMIDE, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
2R9C
 
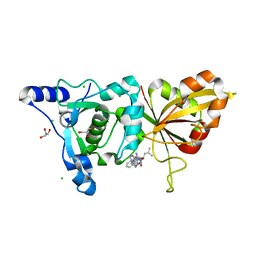 | | Calpain 1 proteolytic core inactivated by ZLAK-3001, an alpha-ketoamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-1 catalytic subunit, ... | | Authors: | Qian, J, Campbell, R.L, Davies, P.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cocrystal structures of primed side-extending alpha-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions.
J.Med.Chem., 51, 2008
|
|
2R9F
 
 | | Calpain 1 proteolytic core inactivated by ZLAK-3002, an alpha-ketoamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-1 catalytic subunit, ... | | Authors: | Qian, J, Campbell, R.L, Davies, P.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cocrystal structures of primed side-extending alpha-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions.
J.Med.Chem., 51, 2008
|
|
2IPP
 
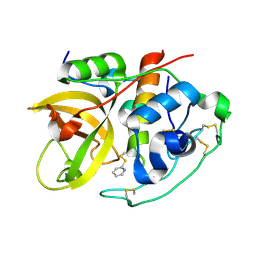 | | Crystal Structure of the tetragonal form of human liver cathepsin B | | Descriptor: | 2-PYRIDINETHIOL, Cathepsin B | | Authors: | Huber, C.P, Campbell, R.L, Hasnain, S, Hirama, T, To, R. | | Deposit date: | 2006-10-12 | | Release date: | 2006-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the tetragonal form of human liver cathepsin B
To be Published
|
|
1I5R
 
 | | TYPE 1 17-BETA HYDROXYSTEROID DEHYDROGENASE EM1745 COMPLEX | | Descriptor: | GLYCEROL, O5'-[9-(3,17B-DIHYDROXY-1,3,5(10)-ESTRATRIEN-16B-YL)-NONANOYL]ADENOSINE, TYPE 1 17 BETA-HYDROXYSTEROID DEHYDROGENASE | | Authors: | Qiu, W, Campbell, R.L, Boivin, P, Poirier, D, Lin, S.-X. | | Deposit date: | 2001-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A concerted, rational design of type 1 17beta-hydroxysteroid dehydrogenase inhibitors:
estradiol-adenosine hybrids with high affinity
FASEB J., 16, 2002
|
|
1IGI
 
 | |
1IGJ
 
 | |
5K8G
 
 | |
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
3DF0
 
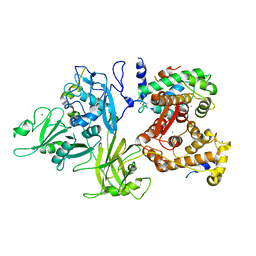 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
5IX9
 
 | | Cell surface anchoring domain | | Descriptor: | Antifreeze protein | | Authors: | Guo, S, Langelaan, D. | | Deposit date: | 2016-03-23 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5J6Y
 
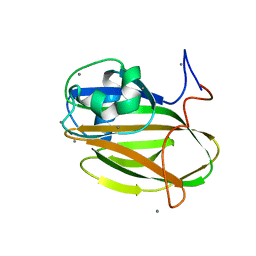 | | Crystal structure of PA14 domain of MpAFP Antifreeze protein | | Descriptor: | Antifreeze protein, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Guo, S. | | Deposit date: | 2016-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5JUH
 
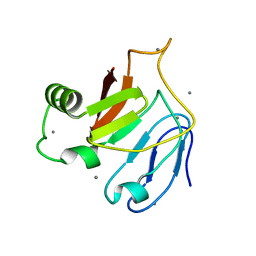 | | Crystal structure of C-terminal domain (RV) of MpAFP | | Descriptor: | Antifreeze protein, CALCIUM ION | | Authors: | Guo, S. | | Deposit date: | 2016-05-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
6W78
 
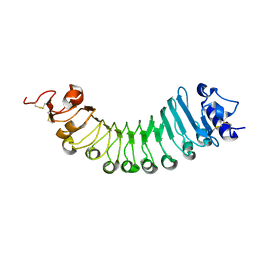 | | crystal structure of a plant ice-binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antifreeze polypeptide | | Authors: | Wang, Y.N, Zhang, H.Q. | | Deposit date: | 2020-03-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Carrot 'antifreeze' protein has an irregular ice-binding site that confers weak freezing point depression but strong inhibition of ice recrystallization.
Biochem.J., 477, 2020
|
|
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
