4MB1
 
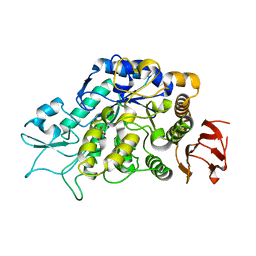 | | The Structure of MalL mutant enzyme G202P from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Oligo-1,6-glucosidase 1 | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4MAZ
 
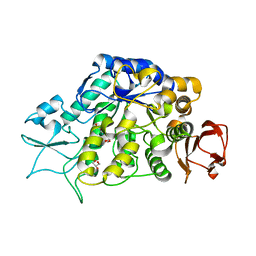 | | The Structure of MalL mutant enzyme V200S from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4M56
 
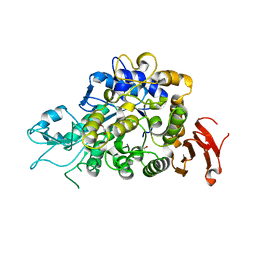 | | The Structure of Wild-type MalL from Bacillus subtilis | | Descriptor: | D-glucose, GLYCEROL, Oligo-1,6-glucosidase 1, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4NOV
 
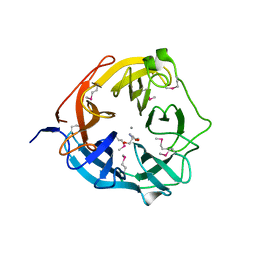 | | Xsa43E, a GH43 family enzyme from Butyrivibrio proteoclasticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Xylosidase/arabinofuranosidase Xsa43E | | Authors: | Till, M, Arcus, V.L. | | Deposit date: | 2013-11-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural analysis of the GH43 enzyme Xsa43E from Butyrivibrio proteoclasticus
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
4M8U
 
 | | The Structure of MalL mutant enzyme V200A from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-13 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
3ITE
 
 | | The third adenylation domain of the fungal SidN non-ribosomal peptide synthetase | | Descriptor: | CHLORIDE ION, SULFATE ION, SidN siderophore synthetase | | Authors: | Lee, T.V, Lott, J.S, Johnson, R.D, Johnson, L.J, Arcus, V.L. | | Deposit date: | 2009-08-28 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis
J.Biol.Chem., 285, 2010
|
|
1RJN
 
 | | The Crystal Structure of MenB (Rv0548c) from Mycobacterium tuberculosis in Complex with the CoA Portion of Naphthoyl CoA | | Descriptor: | 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, COENZYME A, menB | | Authors: | Johnston, J.M, Arcus, V.L, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-19 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of naphthoate synthase (MenB) from Mycobacterium tuberculosis in both native and product-bound forms.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1RJM
 
 | | Crystal Structure of MenB (Rv0548c) from Mycobacterium tuberculosis | | Descriptor: | 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, MenB | | Authors: | Johnston, J.M, Arcus, V.L, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-19 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of naphthoate synthase (MenB) from Mycobacterium tuberculosis in both native and product-bound forms.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1NXJ
 
 | | Structure of Rv3853 from Mycobacterium tuberculosis | | Descriptor: | GLYOXYLIC ACID, L(+)-TARTARIC ACID, Probable S-adenosylmethionine:2-demethylmenaquinone methyltransferase | | Authors: | Johnston, J.M, Arcus, V.L, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-02-10 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Putative Methyltransferase from Mycobacterium tuberculosis: Misannotation of a Genome Clarified by Protein Structural Analysis
J.Bacteriol., 185, 2003
|
|
4E1R
 
 | |
4E1P
 
 | |
4DEV
 
 | |
5U7P
 
 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Trifolium repens | | Descriptor: | Apyrase, PHOSPHATE ION | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
5U7W
 
 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Trifolium repens in complex with adenine and phosphate | | Descriptor: | ADENINE, Apyrase, PHOSPHATE ION | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
5U7X
 
 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Vigna unguiculata subsp. cylindrica (Dolichos biflorus) in complex with phosphate and manganese | | Descriptor: | MANGANESE (II) ION, Nod factor binding lectin-nucleotide phosphohydrolase, PHOSPHATE ION | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
8V5B
 
 | | Structure of the oxygen-insensitive NAD(P)H-dependent nitroreductase NfsB_Ec F70A/F108Y in complex with FMN | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydropteridine reductase, ... | | Authors: | Sharrock, A.V, Ackerley, D.F, Arcus, V. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Evaluation of a Nitroreductase Engineered for Improved Activation of the 5-Nitroimidazole PET Probe SN33623.
Int J Mol Sci, 25, 2024
|
|
7UWT
 
 | | Structure of Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB_Vv F70A/F108Y (NTR 2.0) in complex with FMN at 1.85 Angstroms resolution | | Descriptor: | ACETATE ION, Dihydropteridine reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sharrock, A.V, Arcus, V, Mumm, J.S, Ackerley, D.F. | | Deposit date: | 2022-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Engineered Nitroreductase NTR 2.0 and Impact of F70A and F108Y Substitutions on Substrate Specificity.
Int J Mol Sci, 24, 2023
|
|
3U37
 
 | |
8UCH
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A + ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCE
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCI
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCG
 
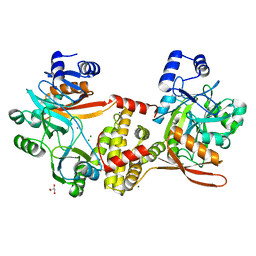 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCF
 
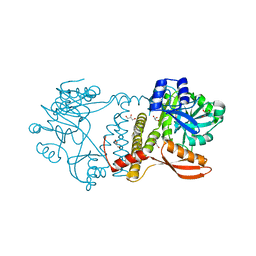 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
4HEC
 
 | |
4HVJ
 
 | |
