3VV3
 
 | | Crystal structure of deseasin MCP-01 from Pseudoalteromonas sp. SM9913 | | Descriptor: | CALCIUM ION, Deseasin MCP-01 | | Authors: | Zhao, G.Y, Gao, X, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2012-07-14 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and mechanistic insights into collagen degradation by a bacterial collagenolytic serine protease in the subtilisin family.
Mol. Microbiol., 90, 2013
|
|
5IQ1
 
 | | Crystal structure of RnTmm mutant Y207S | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5IPY
 
 | | Crystal structure of WT RnTmm | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
6KBW
 
 | | Crystal structure of Tmm from Myroides profundi D25 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethylamine monooxygenase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Oxidation of trimethylamine to trimethylamine N-oxide facilitates high hydrostatic pressure tolerance in a generalist bacterial lineage.
Sci Adv, 7, 2021
|
|
4Z4L
 
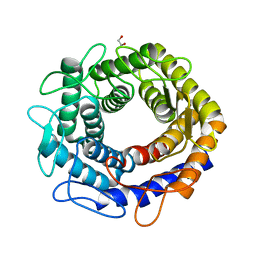 | |
4Z4J
 
 | |
6KCW
 
 | | Structure of alginate lyase Aly36B | | Descriptor: | Alginate lyase, CALCIUM ION, PHOSPHATE ION | | Authors: | Dong, F, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-06-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
4QFL
 
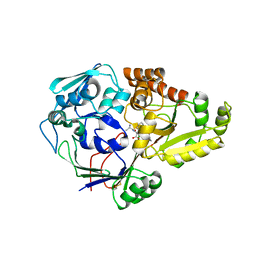 | |
4QFO
 
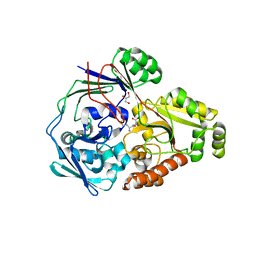 | |
4Q05
 
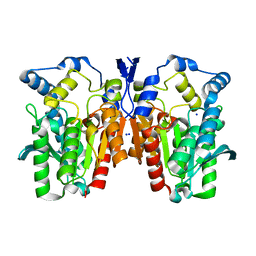 | | Crystal structure of an esterase E25 | | Descriptor: | SODIUM ION, esterase E25 | | Authors: | Li, P.Y, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for dimerization and catalysis of a novel esterase from the GTSAG motif subfamily of the bacterial hormone-sensitive lipase family
J.Biol.Chem., 289, 2014
|
|
4QFK
 
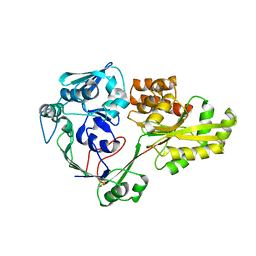 | |
4QFN
 
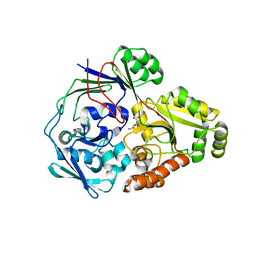 | |
4QFP
 
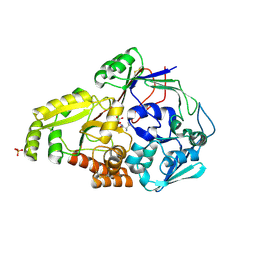 | |
4Q8K
 
 | | Crystal structure of polysaccharide lyase family 18 aly-SJ02 P-CATD | | Descriptor: | Alginase, CALCIUM ION, SULFATE ION | | Authors: | Dong, S, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-04-28 | | Release date: | 2014-09-17 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
4Q8L
 
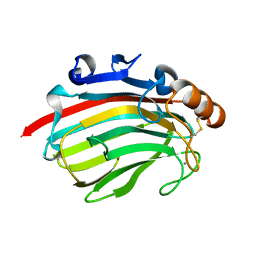 | | Crystal structure of polysacchride lyase family 18 aly-SJ02 r-CATD | | Descriptor: | Alginase, CALCIUM ION | | Authors: | Dong, S, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-04-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
6K7Z
 
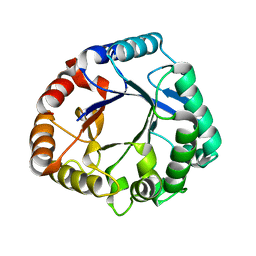 | | Crystal structure of a GH18 chitinase from Pseudoalteromonas aurantia | | Descriptor: | GH18 chiitnase | | Authors: | Wang, Y.J, Li, P.Y, Cao, H.Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Insight Into Chitin Degradation and Thermostability of a Novel Endochitinase From the Glycoside Hydrolase Family 18.
Front Microbiol, 10, 2019
|
|
6LJA
 
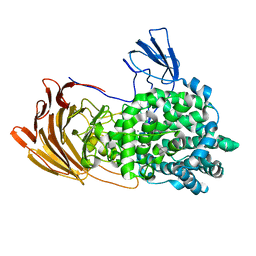 | | Crystal Structure of exoHep from Bacteroides intestinalis DSM 17393 complexed with disaccharide product | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
6LJL
 
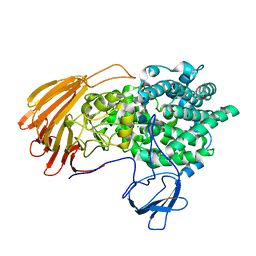 | | Crystal Structure of exoHep-Y390A/H555A complexed with a tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
5XKX
 
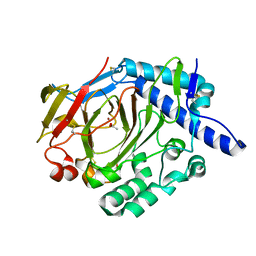 | | Crystal structure of WT DddY | | Descriptor: | ACETATE ION, Uncharacterized protein, ZINC ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
5XKY
 
 | | Crystal structure of DddY Se derivative | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
8HLE
 
 | | Structure of DddY-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, DMSP lyase DddY, ZINC ION | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
5TOU
 
 | | STRUCTURE OF C-PHYCOCYANIN FROM ARCTIC PSEUDANABAENA SP. LW0831 | | Descriptor: | PHYCOCYANOBILIN, Phycocyanin alpha-1 subunit, Phycocyanin beta-1 subunit | | Authors: | Wang, Q.M, Li, C.Y, Su, H.N, Zhang, Y.Z, Xie, B.B. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into the cold adaptation of the photosynthetic pigment-protein C-phycocyanin from an Arctic cyanobacterium
Biochim. Biophys. Acta, 1858, 2017
|
|
2RE9
 
 | | Crystal structure of TL1A at 2.1 A | | Descriptor: | GLYCEROL, MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Guo, F, Kim, S, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1 A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
6KCV
 
 | | Structure of alginate lyase Aly36B mutant K143A/Y185A in complex with alginate tetrasaccharide | | Descriptor: | Alginate lyase, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Dong, F, Zhang, Y.Z, Chen, X.L. | | Deposit date: | 2019-06-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid.
J.Mol.Biol., 431, 2019
|
|
