1GHE
 
 | | CRYSTAL STRUCTURE OF TABTOXIN RESISTANCE PROTEIN COMPLEXED WITH AN ACYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, ACETYLTRANSFERASE | | Authors: | He, H, Ding, Y, Bartlam, M, Sun, F, Le, Y, Qin, X, Tang, H, Zhang, R, Joachimiak, A, Liu, Y, Zhao, N, Rao, Z. | | Deposit date: | 2000-12-13 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Tabtoxin Resistance Protein Complexed with Acetyl
Coenzyme A Reveals the Mechanism for beta-Lactam Acetylation
J.Mol.Biol., 325, 2003
|
|
5J8V
 
 | |
5JQM
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 1.5 Angstroms Resolution | | Descriptor: | Protein UPS1, mitochondrial,Mitochondrial distribution and morphology protein 35 | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
5JQL
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 2.9 Angstroms Resolution | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
3F3H
 
 | | Crystal structure and anti-tumor activity of LZ-8 from the fungus Ganoderma lucidium | | Descriptor: | Immunomodulatory protein Ling Zhi-8 | | Authors: | Zhou, C.Z, Huang, L, He, Y.X, Bao, R, Sun, F, Liang, C, Liu, L. | | Deposit date: | 2008-10-30 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LZ-8 from the medicinal fungus Ganoderma lucidium
Proteins, 75, 2009
|
|
3P2M
 
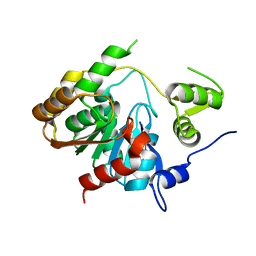 | | Crystal Structure of a Novel Esterase Rv0045c from Mycobacterium tuberculosis | | Descriptor: | POSSIBLE HYDROLASE | | Authors: | Zheng, X.D, Guo, J, Xu, L, Li, H, Zhang, D, Zhang, K, Sun, F, Wen, T, Liu, S, Pang, H. | | Deposit date: | 2010-10-03 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Novel Esterase Rv0045c from Mycobacterium tuberculosis
Plos One, 6, 2011
|
|
4CKH
 
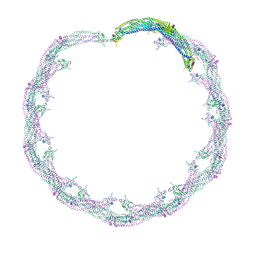 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
4CKG
 
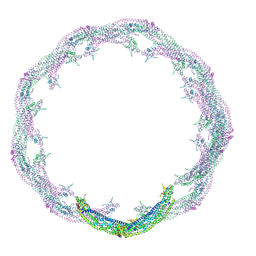 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
2ZGJ
 
 | | Crystal Structure of D86N-GzmM Complexed with Its Optimal Synthesized Substrate | | Descriptor: | Granzyme M, SSGKVPLS, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ZGH
 
 | | Crystal Structure of active granzyme M bound to its product | | Descriptor: | Granzyme M, SSGKVPL, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-22 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ZKS
 
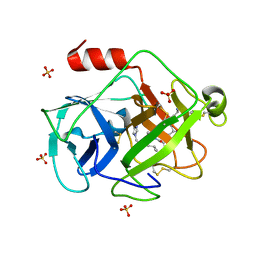 | | Structural insights into the proteolytic machinery of apoptosis-inducing Granzyme M | | Descriptor: | Granzyme M, SULFATE ION, hGzmM inhibitor | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Yang, X, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-03-28 | | Release date: | 2009-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
3A2A
 
 | | The structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1 | | Descriptor: | CHLORIDE ION, Voltage-gated hydrogen channel 1 | | Authors: | Li, S.J, Unno, H, Zhou, Q, Zhao, Q, Zhai, Y, Sun, F. | | Deposit date: | 2009-05-08 | | Release date: | 2010-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1.
J.Biol.Chem., 285, 2010
|
|
2ZGC
 
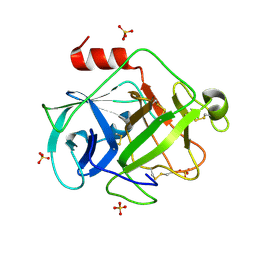 | | Crystal Structure of Active Human Granzyme M | | Descriptor: | Granzyme M, SULFATE ION | | Authors: | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
3IZ3
 
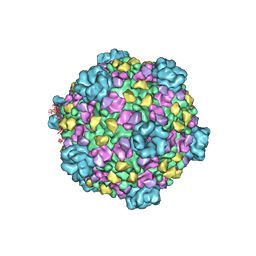 | | CryoEM structure of cytoplasmic polyhedrosis virus | | Descriptor: | Structural protein VP1, Structural protein VP3, Viral structural protein 5 | | Authors: | Cheng, L, Sun, J, Zhang, K, Mou, Z, Huang, X, Ji, G, Sun, F, Zhang, J, Zhu, P. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8H77
 
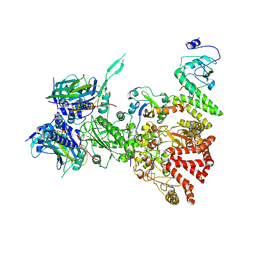 | | Hsp90-AhR-p23-XAP2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
1KU5
 
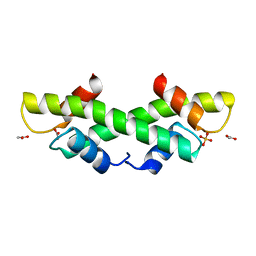 | | Crystal Structure of recombinant histone HPhA from hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, HPhA, SULFATE ION | | Authors: | Li, T, Sun, F, Ji, X, Feng, Y, Rao, Z. | | Deposit date: | 2002-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure based hyperthermostability of archaeal histone HPhA from Pyrococcus horikoshii
J.MOL.BIOL., 325, 2003
|
|
7VOP
 
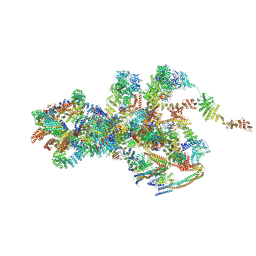 | | Cryo-EM structure of Xenopus laevis nuclear pore complex cytoplasmic ring subunit | | Descriptor: | GATOR complex protein SEC13, IL4I1 protein, MGC154553 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7VCI
 
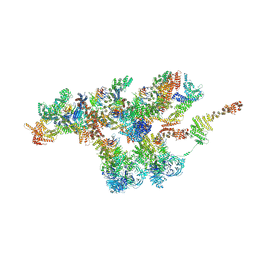 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
3J1P
 
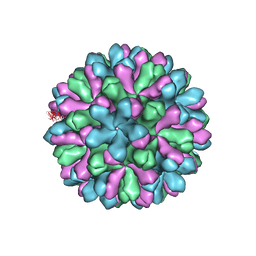 | | Atomic model of rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Liu, Y, Sun, F. | | Deposit date: | 2012-04-09 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
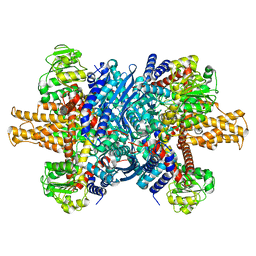
7VDA
 
 | |
7VDC
 
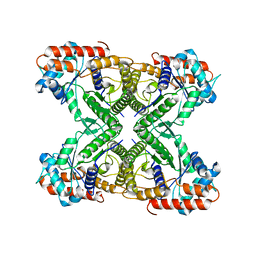 | |
7VD8
 
 | |
7VDF
 
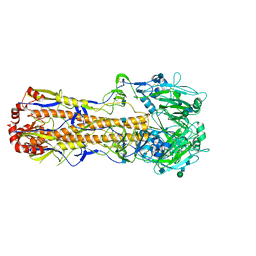 | |
7VD9
 
 | | 2.29 A structure of the human catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, H.C, Zhang, Y, Sun, F. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI.
Nat Commun, 12, 2021
|
|
7VDE
 
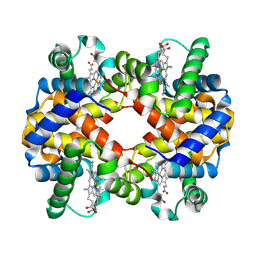 | | 3.6 A structure of the human hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, H.C, Zhang, Y, Sun, F. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI.
Nat Commun, 12, 2021
|
|
