6WSG
 
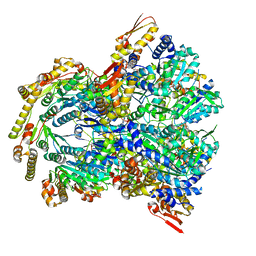 | | ClpX-ClpP complex bound to ssrA-tagged GFP, intermediate complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, Green fluorescent protein, ... | | Authors: | Fei, X, Sauer, R.T. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
3LGV
 
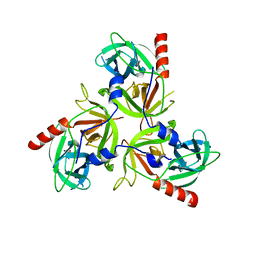 | | H198P mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
3LGW
 
 | |
3LGI
 
 | |
3LGT
 
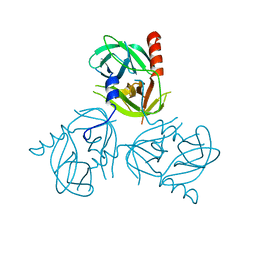 | |
3LH3
 
 | | DFP modified DegS delta PDZ | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
5TXR
 
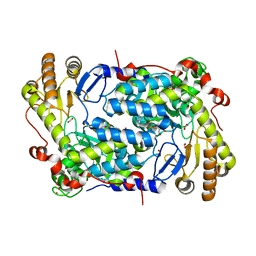 | | Structure of ALAS from S. cerevisiae non-covalently bound to PLP cofactor | | Descriptor: | 5-aminolevulinate synthase, mitochondrial, FORMIC ACID, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
5TXV
 
 | | HslU P21 cell with 4 hexamers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU | | Authors: | Grant, R.A, Chen, J, Glynn, S.E, Sauer, R.T. | | Deposit date: | 2016-11-17 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (7.086 Å) | | Cite: | Covalently linked HslU hexamers support a probabilistic mechanism that links ATP hydrolysis to protein unfolding and translocation.
J. Biol. Chem., 292, 2017
|
|
5TXT
 
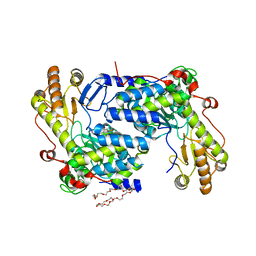 | | Structure of asymmetric apo/holo ALAS dimer from S. cerevisiae | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 5-aminolevulinate synthase, mitochondrial, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
4YKA
 
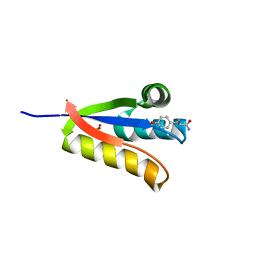 | | The structure of Agrobacterium tumefaciens ClpS2 in complex with L-tyrosinamide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS 2, L-TYROSINAMIDE, SULFATE ION | | Authors: | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
4YJX
 
 | | The structure of Agrobacterium tumefaciens ClpS2 bound to L-phenylalaninamide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS 2, PHENYLALANINE AMIDE, SULFATE ION | | Authors: | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2015-03-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
4YJM
 
 | |
3B8J
 
 | |
4I5O
 
 | | Crystal Structure of W-W-R ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.4787 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I9K
 
 | | Crystal structure of symmetric W-W-W ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-12-05 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.0003 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I63
 
 | | Crystal Structure of E-R ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.709 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I34
 
 | | Crystal Structure of W-W-W ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.1218 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I81
 
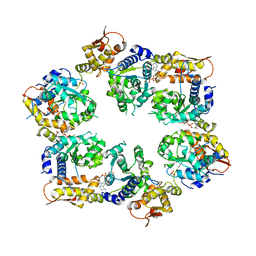 | | Crystal Structure of ATPgS bound ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-12-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8182 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I4L
 
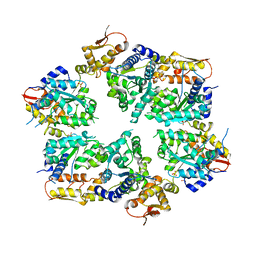 | | Crystal Structure of Nucleotide-Bound W-W-W ClpX Hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6981 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
2HDD
 
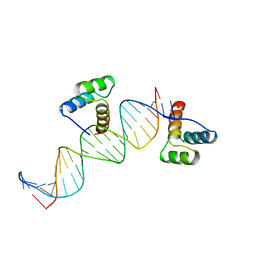 | | ENGRAILED HOMEODOMAIN Q50K VARIANT DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*GP*GP*GP*GP*AP*TP*TP*AP*CP*AP*TP*GP*G P*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*CP*CP*C P*CP*GP*GP*A)-3'), PROTEIN (ENGRAILED HOMEODOMAIN Q50K) | | Authors: | Tucker-Kellogg, L, Rould, M.A, Chambers, K.A, Ades, S.E, Sauer, R.T, Pabo, C.O. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engrailed (Gln50-->Lys) homeodomain-DNA complex at 1.9 A resolution: structural basis for enhanced affinity and altered specificity.
Structure, 5, 1997
|
|
5JI3
 
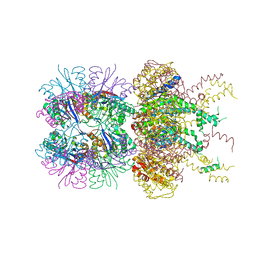 | | HslUV complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
5JI2
 
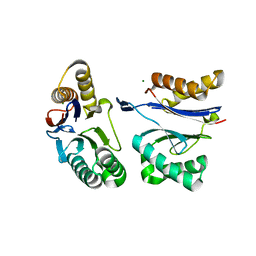 | | HslU L199Q in HslUV complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
4U0G
 
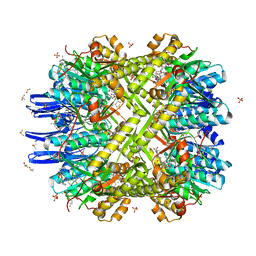 | | Crystal Structure of M. tuberculosis ClpP1P2 bound to ADEP and agonist | | Descriptor: | ADEP-2B5Me, ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, ... | | Authors: | Schmitz, K.R, Carney, D.W, Sello, J.K, Sauer, R.T. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1978 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U0H
 
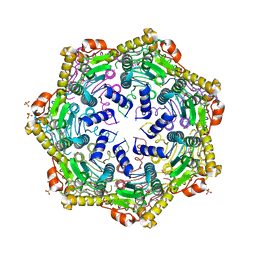 | | Crystal Structure of M. tuberculosis ClpP1P1 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, SULFATE ION | | Authors: | Schmitz, K.R, Carney, D.W, Sello, J.K, Sauer, R.T. | | Deposit date: | 2014-07-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2479 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3GCN
 
 | |
