1WLG
 
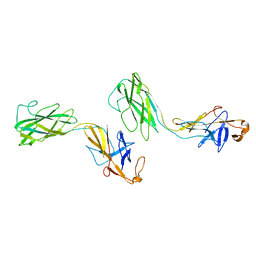 | | Crystal structure of FlgE31, a major fragment of the hook protein | | Descriptor: | Flagellar hook protein flgE | | Authors: | Samatey, F.A, Matsunami, H, Imada, K, Nagashima, S, Shaikh, T.R, Thomas, D.R, DeRosier, D.J, Kitao, A, Namba, K. | | Deposit date: | 2004-06-25 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism.
Nature, 431, 2004
|
|
8K6J
 
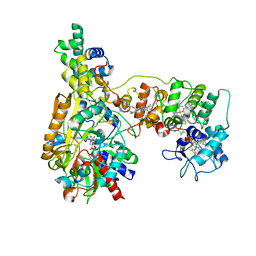 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-H1147A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (H1147A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8K6K
 
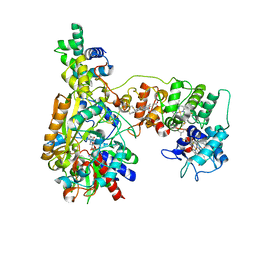 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (N1146A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
6LU1
 
 | | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Gerle, C, Kawamoto, A, Kato, T, Namba, K, Nowaczyk, M.M, Rogner, M, Misumi, Y, Frank, A, Eithar, E.M. | | Deposit date: | 2020-01-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
3A5I
 
 | |
3J0R
 
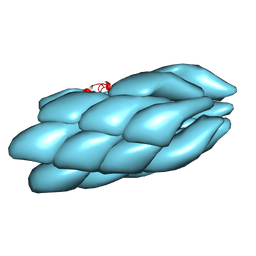 | | Model of a type III secretion system needle based on a 7 Angstrom resolution cryoEM map | | Descriptor: | Protein mxiH | | Authors: | Fujii, T, Cheung, M, Blanco, A, Kato, T, Blocker, A.J, Namba, K. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of a type III secretion needle at 7-A resolution provides insights into its assembly and signaling mechanisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3AJW
 
 | | Structure of FliJ, a soluble component of flagellar type III export apparatus | | Descriptor: | Flagellar fliJ protein, MERCURY (II) ION | | Authors: | Imada, K, Ibuki, T, Minamino, T, Namba, K. | | Deposit date: | 2010-06-23 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases
Nat.Struct.Mol.Biol., 18, 2011
|
|
8IBN
 
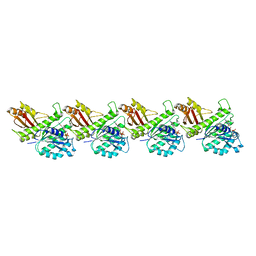 | | Cryo-EM structure of KpFtsZ single filament | | Descriptor: | Cell division protein FtsZ, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, POTASSIUM ION | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hibino, K, Konishi, T, Kato, Y, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
3J82
 
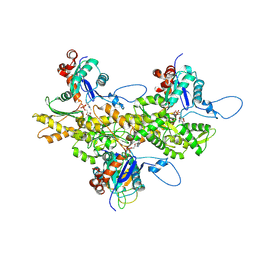 | | Electron cryo-microscopy of DNGR-1 in complex with F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Hanc, P, Fujii, T, Yamada, Y, Huotari, J, Schulz, O, Ahrens, S, Kjaer, S, Way, M, Namba, K, Reis e Sousa, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-05-20 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens.
Immunity, 42, 2015
|
|
8H1O
 
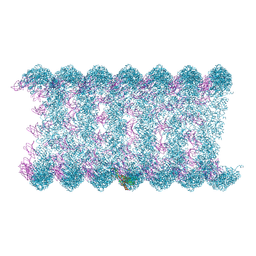 | | Cryo-EM structure of KpFtsZ-monobody double helical tube | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Mb(Ec/KpFtsZ_S1) | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
8ZDU
 
 | | Structure of the RBM3 ring of Salmonella flagellar MS-ring protein FliF with C34 symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Kinoshita, M, Makino, F, Miyata, T, Imada, K, Minamino, T, Namba, K. | | Deposit date: | 2024-05-03 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for assembly and function of the Salmonella flagellar MS-ring with three different symmetries.
Commun Biol, 8, 2025
|
|
8ZDS
 
 | | Structure of the Salmonella flagellar MS-ring with C11 symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Kinoshita, M, Makino, F, Miyata, T, Imada, K, Minamino, T, Namba, K. | | Deposit date: | 2024-05-03 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for assembly and function of the Salmonella flagellar MS-ring with three different symmetries.
Commun Biol, 8, 2025
|
|
8ZDT
 
 | | Structure of the RBM3 ring of Salmonella flagellar MS-ring protein FliF with C33 symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Kinoshita, M, Makino, F, Miyata, T, Imada, K, Minamino, T, Namba, K. | | Deposit date: | 2024-05-03 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for assembly and function of the Salmonella flagellar MS-ring with three different symmetries.
Commun Biol, 8, 2025
|
|
1IO1
 
 | | CRYSTAL STRUCTURE OF F41 FRAGMENT OF FLAGELLIN | | Descriptor: | PHASE 1 FLAGELLIN | | Authors: | Samatey, F.A, Imada, K, Nagashima, S, Vondervisz, F, Kumasaka, T, Yamamoto, M, Namba, K. | | Deposit date: | 2000-12-28 | | Release date: | 2001-04-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling
Nature, 410, 2001
|
|
8GY2
 
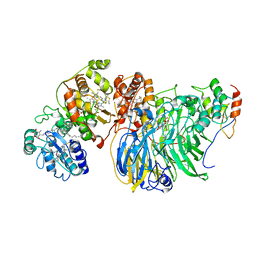 | | Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans | | Descriptor: | Alcohol dehydrogenase (quinone), cytochrome c subunit, dehydrogenase subunit, ... | | Authors: | Adachi, T, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Acs Catalysis, 13, 2023
|
|
8GY3
 
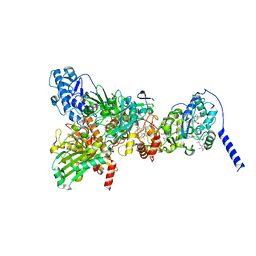 | | Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), Cytochrome c subunit of aldehyde dehydrogenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Adachi, T, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Acs Catalysis, 13, 2023
|
|
3VJP
 
 | |
3TEE
 
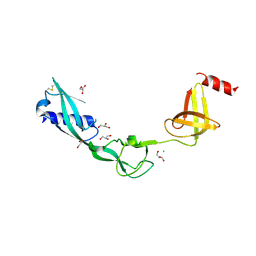 | | Crystal Structure of Salmonella FlgA in open form | | Descriptor: | CHLORIDE ION, Flagella basal body P-ring formation protein flgA, GLYCEROL | | Authors: | Matsunami, H, Samatey, F.A, Namba, K. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica
Sci Rep, 6, 2016
|
|
3VKI
 
 | |
1GTT
 
 | | CRYSTAL STRUCTURE OF HPCE | | Descriptor: | 4-HYDROXYPHENYLACETATE DEGRADATION BIFUNCTIONAL ISOMERASE/DECARBOXYLASE, CALCIUM ION | | Authors: | Tame, J.R.H, Namba, K, Dodson, E.J, Roper, D.I. | | Deposit date: | 2002-01-18 | | Release date: | 2002-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Hpce, a Bifunctional Decarboxylase/Isomerase with a Multifunctional Fold.
Biochemistry, 41, 2002
|
|
4A6J
 
 | | Structural model of ParM filament based on CryoEM map | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | Gayathri, P, Fujii, T, Moller-Jensen, J, Van Den Ent, F, Namba, K, Lowe, J. | | Deposit date: | 2011-11-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
1I7O
 
 | | CRYSTAL STRUCTURE OF HPCE | | Descriptor: | 4-HYDROXYPHENYLACETATE DEGRADATION BIFUNCTIONAL ISOMERASE/DECARBOXYLASE, CALCIUM ION | | Authors: | Tame, J.R.H, Namba, K, Dodson, E.J, Roper, D.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-03-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of HpcE, a multi-functional enzyme fold
To be Published
|
|
2D4V
 
 | | Crystal structure of NAD dependent isocitrate dehydrogenase from Acidithiobacillus thiooxidans | | Descriptor: | CITRATE ANION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, isocitrate dehydrogenase | | Authors: | Imada, K, Tamura, T, Namba, K, Inagaki, K. | | Deposit date: | 2005-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity
Proteins, 70, 2008
|
|
