2VSQ
 
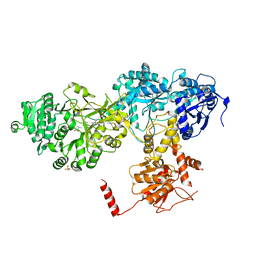 | | Structure of surfactin A synthetase C (SrfA-C), a nonribosomal peptide synthetase termination module | | Descriptor: | LEUCINE, SULFATE ION, SURFACTIN SYNTHETASE SUBUNIT 3 | | Authors: | Tanovic, A, Samel, S.A, Essen, L.-O, Marahiel, M.A. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the termination module of a nonribosomal peptide synthetase.
Science, 321, 2008
|
|
2XUZ
 
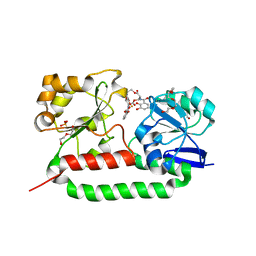 | | Crystal structure of the triscatecholate siderophore binding protein FeuA from Bacillus subtilis complexed with Ferri-Enterobactin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, IRON-UPTAKE SYSTEM-BINDING PROTEIN, ... | | Authors: | Peuckert, F, Miethke, M, Schwoerer, C.J, Albrecht, A.G, Oberthuer, M, Marahiel, M.A. | | Deposit date: | 2010-10-22 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Siderophore Binding Protein Feua Shows Limited Promiscuity Toward Exogenous Triscatecholates
Chem.Biol., 18, 2011
|
|
2WBP
 
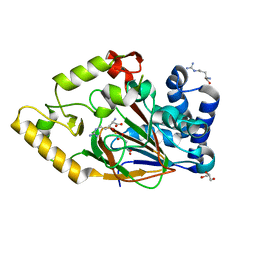 | | Crystal structure of VioC in complex with Fe(II), (2S,3S)- hydroxyarginine, and succinate | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, ARGININE, FE (II) ION, ... | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
2WBQ
 
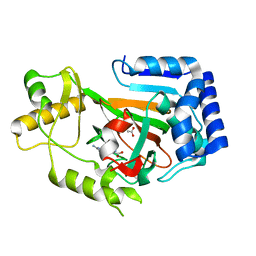 | | Crystal structure of VioC in complex with (2S,3S)-hydroxyarginine | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, ACETATE ION, L-ARGININE BETA-HYDROXYLASE | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
2XV1
 
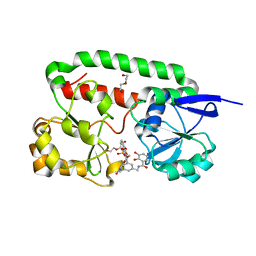 | | Crystal structure of the triscatecholate siderophore binding protein FeuA from Bacillus subtilis complexed with Ferric MECAM | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, IRON-UPTAKE SYSTEM-BINDING PROTEIN, ... | | Authors: | Peuckert, F, Miethke, M, Schwoerer, C.J, Albrecht, A.G, Oberthuer, M, Marahiel, M.A. | | Deposit date: | 2010-10-22 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Siderophore Binding Protein Feua Shows Limited Promiscuity Toward Exogenous Triscatecholates
Chem.Biol., 18, 2011
|
|
2JGP
 
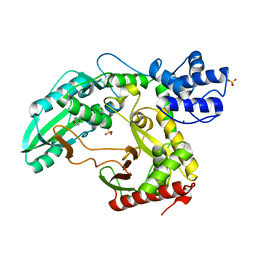 | | Structure of the TycC5-6 PCP-C bidomain of the tyrocidine synthetase TycC | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, SULFATE ION, ... | | Authors: | Samel, S.A, Schoenafinger, G, Knappe, T.A, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2007-02-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Insights Into a Peptide Bond-Forming Bidomain from a Nonribosomal Peptide Synthetase.
Structure, 15, 2007
|
|
2LX6
 
 | |
2M37
 
 | | Structure of lasso peptide astexin-1 | | Descriptor: | ASTEXIN-1 | | Authors: | Zimmermann, M, Hegemann, J.D, Xie, X, Marahiel, M.A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | The astexin-1 lasso peptides: biosynthesis, stability, and structural studies.
Chem.Biol., 20, 2013
|
|
2K2Q
 
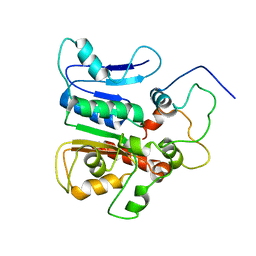 | | complex structure of the external thioesterase of the Surfactin-synthetase with a carrier domain | | Descriptor: | Surfactin synthetase thioesterase subunit, Tyrocidine synthetase 3 (Tyrocidine synthetase III) | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guntert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Dotsch, V. | | Deposit date: | 2008-04-10 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
Nature, 454, 2008
|
|
2MLJ
 
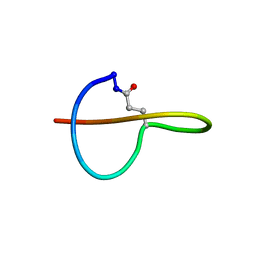 | |
2MFV
 
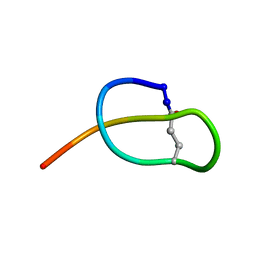 | |
2MMW
 
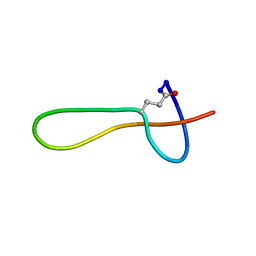 | |
2MMT
 
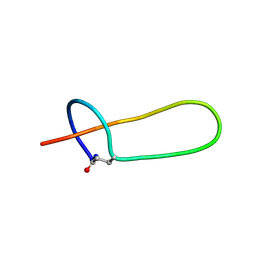 | | Lasso peptide-based integrin inhibitor: Microcin J25 variant with RGDF substitution of Gly12-Ile13-Gly14-Thr15 | | Descriptor: | Microcin J25 RGDF mutant | | Authors: | Hegemann, J.D, Zimmermann, M, Knappe, T.A, Xie, X, Marahiel, M.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-07-02 | | Last modified: | 2014-07-23 | | Method: | SOLUTION NMR | | Cite: | Rational improvement of the affinity and selectivity of integrin binding of grafted lasso peptides.
J.Med.Chem., 57, 2014
|
|
5OQZ
 
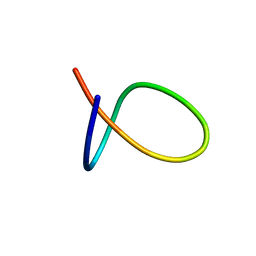 | |
1QR0
 
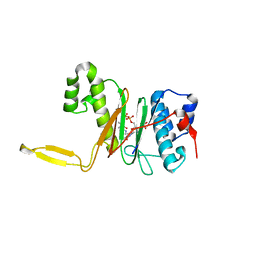 | | CRYSTAL STRUCTURE OF THE 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP-COENZYME A COMPLEX | | Descriptor: | 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP, COENZYME A, MAGNESIUM ION | | Authors: | Reuter, K, Mofid, R.M, Marahiel, A.M, Ficner, R. | | Deposit date: | 1999-06-17 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily.
EMBO J., 18, 1999
|
|
1CSQ
 
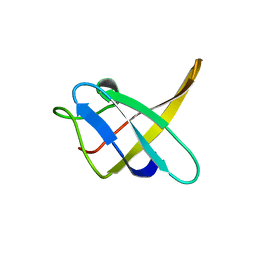 | |
1CSP
 
 | |
1NMG
 
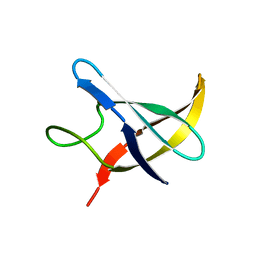 | |
1NMF
 
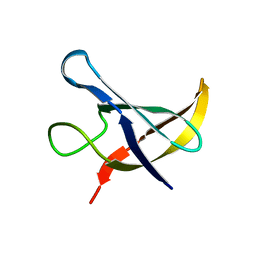 | |
4R1J
 
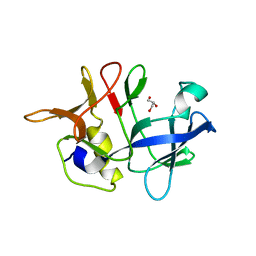 | | Crystal structure of Arc1p-C | | Descriptor: | GLYCEROL, GU4 nucleic-binding protein 1 | | Authors: | Altegoer, F, Bange, G. | | Deposit date: | 2014-08-06 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Synthetic Adenylation-Domain-Based tRNA-Aminoacylation Catalyst.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5T3E
 
 | |
5DKJ
 
 | | Yeast 20S proteasome in complex with octreotide-PI | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Beck, P, Cui, H, Groll, M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeted Delivery of Proteasome Inhibitors to Somatostatin-Receptor-Expressing Cancer Cells by Octreotide Conjugation.
Chemmedchem, 10, 2015
|
|
5F2V
 
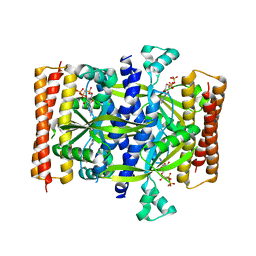 | | Crystal structure of the small alarmone synthethase 1 from Bacillus subtilis bound to AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GTP pyrophosphokinase YjbM, MAGNESIUM ION | | Authors: | Steinchen, W, Schuhmacher, J.S, Altegoer, F, Bange, G. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5J8Q
 
 | |
5DEC
 
 | |
