2IMF
 
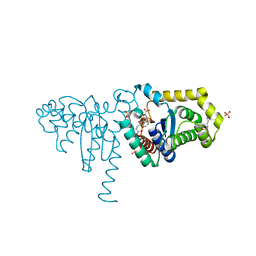 | | 2-Hydroxychromene-2-carboxylate Isomerase: a Kappa Class Glutathione-S-Transferase from Pseudomonas putida | | Descriptor: | 2-hydroxychromene-2-carboxylate isomerase, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, 4-(2-METHOXYPHENYL)-2-OXOBUT-3-ENOIC ACID, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic acid isomerase: a kappa class glutathione transferase from Pseudomonas putida.
Biochemistry, 46, 2007
|
|
2IME
 
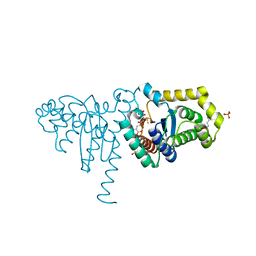 | | 2-Hydroxychromene-2-carboxylate Isomerase: a Kappa Class Glutathione-S-Transferase from Pseudomonas putida | | Descriptor: | (2S)-2-HYDROXY-2H-CHROMENE-2-CARBOXYLIC ACID, (3E)-4-(2-HYDROXYPHENYL)-2-OXOBUT-3-ENOIC ACID, 2-hydroxychromene-2-carboxylate isomerase, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic Acid Isomerase: A Kappa Class Glutathione Transferase from Pseudomonas putida
Biochemistry, 46, 2007
|
|
1GSQ
 
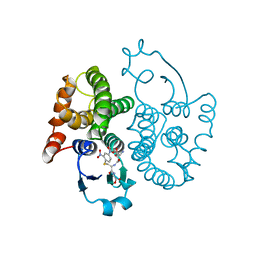 | | THREE-DIMENSIONAL STRUCTURE, CATALYTIC PROPERTIES AND EVOLUTION OF A SIGMA CLASS GLUTATHIONE TRANSFERASE FROM SQUID, A PROGENITOR OF THE LENS-CRYSTALLINS OF CEPHALOPODS | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Ji, X, Rosenvinge, E.C.V, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1995-01-09 | | Release date: | 1995-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure, catalytic properties, and evolution of a sigma class glutathione transferase from squid, a progenitor of the lens S-crystallins of cephalopods.
Biochemistry, 34, 1995
|
|
3R3E
 
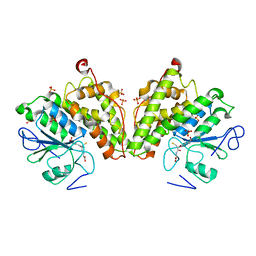 | | The glutathione bound structure of YqjG, a glutathione transferase homolog from Escherichia coli K-12 | | Descriptor: | GLUTATHIONE, SULFATE ION, Uncharacterized protein yqjG | | Authors: | Branch, M.C, Cook, P.D, Harp, J.M, Armstrong, R.N. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Crystal Structure Analysis of yqjG from Escherichia Coli K-12
To be Published
|
|
3R2Q
 
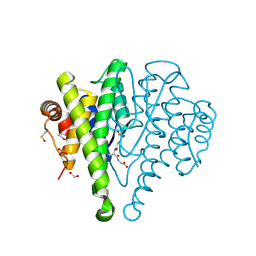 | | Crystal Structure Analysis of yibF from E. Coli | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, PHOSPHATE ION, ... | | Authors: | Ladner, J.E, Harp, J, Schaab, M, Stournan, N.V, Armstrong, R.N. | | Deposit date: | 2011-03-14 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural and Functional Genomics of YibF, a Glutathione Transferase Homologue from Escherichia coli
to be published
|
|
4PX1
 
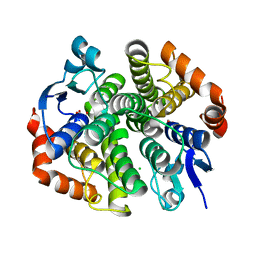 | | CRYSTAL STRUCTURE OF Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE (TARGET EFI-507068) | | Descriptor: | CHLORIDE ION, MALONIC ACID, Maleylacetoacetate isomerase (Glutathione S-transferase) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4PXO
 
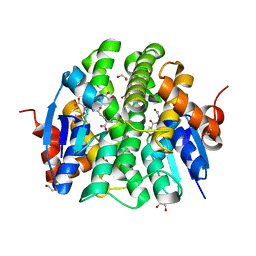 | | Crystal structure of Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE AND GSH (TARGET EFI-507068) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, MALONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-24 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from Methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
1N2A
 
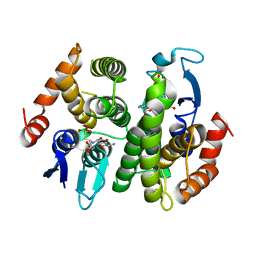 | | Crystal Structure of a Bacterial Glutathione Transferase from Escherichia coli with Glutathione Sulfonate in the Active Site | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase | | Authors: | Rife, C.L, Parsons, J.F, Xiao, G, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2002-10-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved structural elements in glutathione transferase homologues encoded in the genome of Escherichia coli
Proteins, 53, 2003
|
|
1MTC
 
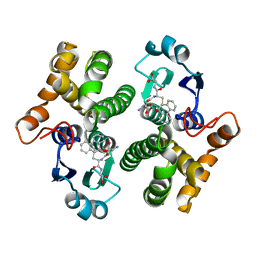 | | GLUTATHIONE TRANSFERASE MUTANT Y115F | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, Glutathione S-transferase YB1 | | Authors: | Ladner, J.E, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Local protein dynamics and catalysis: detection of segmental motion associated with rate-limiting product release by a glutathione transferase
Biochemistry, 41, 2002
|
|
1NKI
 
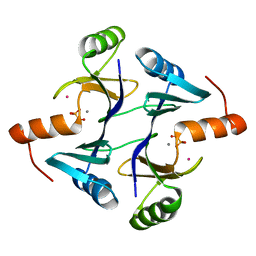 | | CRYSTAL STRUCTURE OF THE FOSFOMYCIN RESISTANCE PROTEIN A (FOSA) CONTAINING BOUND PHOSPHONOFORMATE | | Descriptor: | MANGANESE (II) ION, PHOSPHONOFORMIC ACID, POTASSIUM ION, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2003-01-03 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Phosphonoformate: a minimal transition state analogue inhibitor of the fosfomycin resistance protein, FosA.
Biochemistry, 43, 2004
|
|
1NPB
 
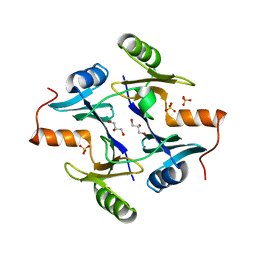 | | Crystal structure of the fosfomycin resistance protein from transposon Tn2921 | | Descriptor: | GLYCEROL, SULFATE ION, fosfomycin-resistance protein | | Authors: | Pakhomova, S, Rife, C.L, Armstrong, R.N, Newcomer, M.E. | | Deposit date: | 2003-01-17 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of fosfomycin resistance protein FosA from transposon Tn2921.
Protein Sci., 13, 2004
|
|
1NNR
 
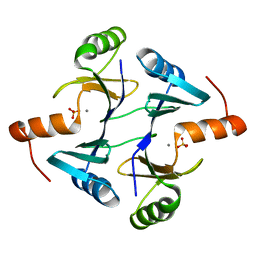 | | Crystal structure of a probable fosfomycin resistance protein (PA1129) from Pseudomonas aeruginosa with sulfate present in the active site | | Descriptor: | MANGANESE (II) ION, SULFATE ION, probable fosfomycin resistance protein | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2003-01-14 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Phosphonoformate: a minimal transition state analogue inhibitor of the fosfomycin resistance protein, FosA.
Biochemistry, 43, 2004
|
|
3UAP
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501774) from methylococcus capsulatus str. bath | | Descriptor: | GLYCEROL, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Methylococcus Capsulatus
To be Published
|
|
3UBK
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutathione transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
3UAR
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501774) from methylococcus capsulatus str. bath with gsh bound | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Methylococcus Capsulatus
To be Published
|
|
3UBL
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans with gsh bound | | Descriptor: | CHLORIDE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
3TOT
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE (TARGET EFI-501058) from Ralstonia solanacearum GMI1000 | | Descriptor: | ACETATE ION, Glutathione s-transferase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of GLUTATHIONE S-TRANSFERASE from Ralstonia solanacearum
To be Published
|
|
3TOU
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE (TARGET EFI-501058) from Ralstonia solanacearum GMI1000 with GSH bound | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione s-transferase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of GLUTATHIONE S-TRANSFERASE from Ralstonia solanacearum
To be Published
|
|
1LQK
 
 | | High Resolution Structure of Fosfomycin Resistance Protein A (FosA) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2002-05-10 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 A resolution by MAD
phasing off the L-III edge of Tl(+)
J.Am.Chem.Soc., 124, 2002
|
|
1LQO
 
 | | Crystal Strutcure of the Fosfomycin Resistance Protein A (FosA) Containing Bound Thallium Cations | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROBABLE Fosfomycin Resistance Protein, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2002-05-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 A resolution by MAD
phasing off the L-III edge of Tl(+)
J.Am.Chem.Soc., 124, 2002
|
|
1LQP
 
 | | CRYSTAL STRUCTURE OF THE FOSFOMYCIN RESISTANCE PROTEIN (FOSA) CONTAINING BOUND SUBSTRATE | | Descriptor: | FOSFOMYCIN, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2002-05-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 A resolution by MAD
phasing off the L-III edge of Tl(+)
J.Am.Chem.Soc., 124, 2002
|
|
1R4W
 
 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
1R9C
 
 | | Crystal Structure of Fosfomycin Resistance Protein FosX from Mesorhizobium Loti | | Descriptor: | MANGANESE (II) ION, glutathione transferase | | Authors: | Fillgrove, K.L, Pakhomova, S, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2003-10-28 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms.
J.Am.Chem.Soc., 125, 2003
|
|
2P7M
 
 | | Crystal structure of monoclinic form of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes at pH 6.5 | | Descriptor: | Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
2P7L
 
 | | Crystal structure of monoclinic form of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes at pH 5.75 | | Descriptor: | GLYCEROL, Glyoxalase family protein | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
