1K34
 
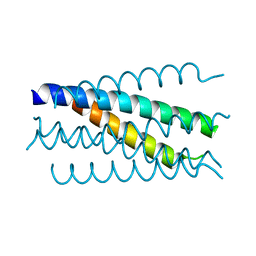 | | Crystal structure analysis of gp41 core mutant | | Descriptor: | Transmembrane glycoprotein GP41 | | Authors: | Shu, W, Lu, M. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion.
Biochemistry, 41, 2002
|
|
2POJ
 
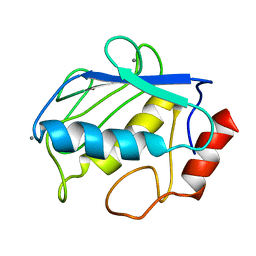 | |
1LDA
 
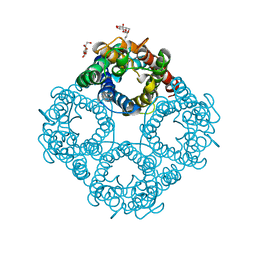 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LDI
 
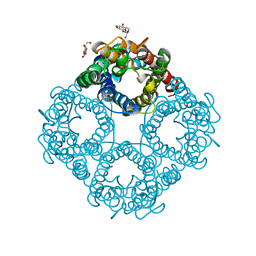 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
2RC5
 
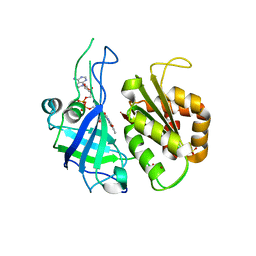 | | Refined structure of FNR from Leptospira interrogans | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase, SULFATE ION, ... | | Authors: | Nascimento, A.S, Catalano-Dupuy, D.L, Polikarpov, I, Ceccarelli, E.A. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | Crystal structures of Leptospira interrogans FAD-containing ferredoxin-NADP+ reductase and its complex with NADP+.
Bmc Struct.Biol., 7, 2007
|
|
2RC6
 
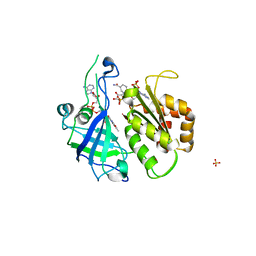 | | Refined structure of FNR from Leptospira interrogans bound to NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Nascimento, A.S, Catalano-Dupuy, D.L, Ceccarelli, E.A, Polikarpov, I. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Leptospira interrogans FAD-containing ferredoxin-NADP+ reductase and its complex with NADP+.
Bmc Struct.Biol., 7, 2007
|
|
5OV7
 
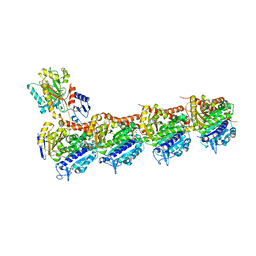 | | tubulin - rigosertib complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Menchon, G, Prota, A.E, Steinmetz, M, Jost, M. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent.
Mol. Cell, 68, 2017
|
|
5NLX
 
 | | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5O7M
 
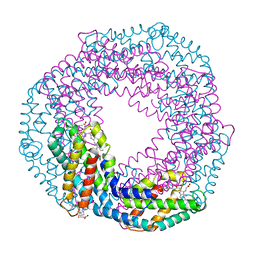 | | Single-shot pink beam serial crystallography: Phycocyanin (One chip, chip_1) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2017-06-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5O5W
 
 | | Molybdenum storage protein room-temperature structure determined by serial millisecond crystallography | | Descriptor: | (mu3-oxo)-tris(mu2-oxo)-nonakisoxo-trimolybdenum (VI), ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Steffen, B, Weinert, T, Ermler, U, Standfuss, J. | | Deposit date: | 2017-06-02 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
3INK
 
 | |
5NJM
 
 | | Lysozyme room-temperature structure determined by serial millisecond crystallography | | Descriptor: | Lysozyme C | | Authors: | Weinert, T, Vera, L, Marsh, M, James, D, Gashi, D, Nogly, P, Jaeger, K, Standfuss, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NJQ
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 4ET8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NJP
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 1HEW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
8RKG
 
 | | Crystal structure of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICINE, ... | | Authors: | Nishio, S, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
6R2O
 
 | | Hemoglobin structure from serial crystallography with a 3D-printed nozzle. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Hemoglobin subunit alpha, ... | | Authors: | Oberthuer, D, Yefanov, O, Sarrou, I. | | Deposit date: | 2019-03-18 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Ultracompact 3D microfluidics for time-resolved structural biology.
Nat Commun, 11, 2020
|
|
1CPB
 
 | |
1E07
 
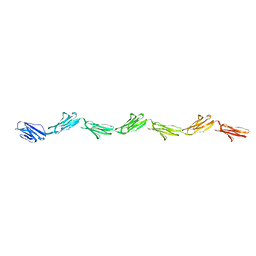 | |
2K37
 
 | | CsmA | | Descriptor: | Chlorosome Protein A | | Authors: | Pedersen, M, Dittmer, J, Underhaug, J, Miller, M, Nielsen, N.C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of CsmA: A small antenna protein from the green sulfur bacterium Chlorobium tepidum.
Febs Lett., 582, 2008
|
|
2KYU
 
 | | The solution structure of the PHD3 finger of MLL | | Descriptor: | Histone-lysine N-methyltransferase MLL, ZINC ION | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-08 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression.
Biochemistry, 49, 2010
|
|
2L4W
 
 | | NMR structure of the Xanthomonas VirB7 | | Descriptor: | Uncharacterized protein | | Authors: | Souza, D.P, Farah, C.S, Salinas, R.K. | | Deposit date: | 2010-10-15 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Component of the Xanthomonadaceae Type IV Secretion System Combines a VirB7 Motif with a N0 Domain Found in Outer Membrane Transport Proteins.
Plos Pathog., 7, 2011
|
|
2KYX
 
 | | Solution structure of the RRM domain of CYP33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression .
Biochemistry, 49, 2010
|
|
2LIE
 
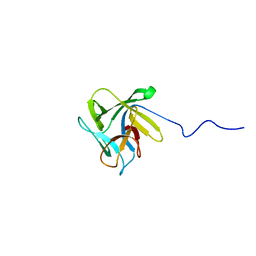 | | NMR structure of the lectin CCL2 | | Descriptor: | CCL2 lectin | | Authors: | Schubert, M, Walti, M.A, Egloff, P, Bleuler-Martinez, S, Aebi, M, Allain, F.F.-H, Kunzler, M. | | Deposit date: | 2011-08-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2LIQ
 
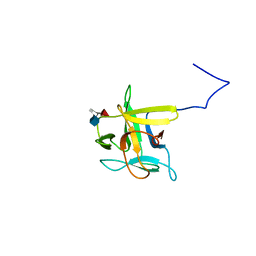 | | Solution structure of CCL2 in complex with glycan | | Descriptor: | CCL2 lectin, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Schubert, M, Bleuler-Martinez, S, Walti, M.A, Egloff, P, Aebi, M, Kuenzler, M, Allain, F.H.-T. | | Deposit date: | 2011-08-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
4R8T
 
 | | Structure of JEV protease | | Descriptor: | CHLORIDE ION, NS3, Serine protease subunit NS2B | | Authors: | Nair, D.T, Weinert, T, Wang, M, Olieric, V. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
