6UG6
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, C3-1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
8T0B
 
 | |
8T1N
 
 | | Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Cascio, D, Sawaya, M.R, Cannon, K.A, Rodriguez, J.A, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8T1M
 
 | | Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Agdanowski, M.P, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6UGB
 
 | | C3 symmetric peptide design number 2, Baby Basil | | Descriptor: | C3 symmetric peptide design number 2, Baby Basil, CHLORIDE ION, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDW
 
 | | S2 symmetric peptide design number 3 crystal form 2, Lurch | | Descriptor: | S2-3, Lurch crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF7
 
 | | S2 symmetric peptide design number 5, Uncle Fester | | Descriptor: | S2-5, Uncle Fester | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF8
 
 | | S2 symmetric peptide design number 6, London Bridge | | Descriptor: | S2-6, London Bridge | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDR
 
 | | S2 symmetric peptide design number 3 crystal form 1, Lurch | | Descriptor: | S2-3, Lurch crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UFU
 
 | | C2 symmetric peptide design number 1, Zappy, crystal form 1 | | Descriptor: | C2-1, Zappy, crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG3
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 1 | | Descriptor: | C3-1, Sporty, crystal form 1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UGC
 
 | | C3 symmetric peptide design number 3 | | Descriptor: | C3-3 cyclic peptide design, CADMIUM ION, SODIUM ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
8UMP
 
 | | T33-ml35 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml35-redesigned-CutA-fold, T33-ml35-redesigned-TPR-domain-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-18 | | Release date: | 2023-11-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
8UF0
 
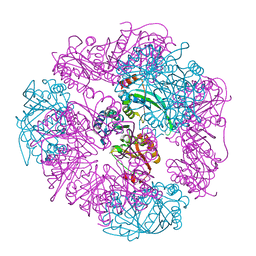 | | T33-ml23 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml23-redesigned-CutA-fold, T33-ml23-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-03 | | Release date: | 2023-11-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
6UUR
 
 | | Human prion protein fibril, M129 variant | | Descriptor: | Major prion protein | | Authors: | Glynn, C, Sawaya, M.R, Ge, P, Zhou, Z.H, Rodriguez, J.A. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of a human prion fibril with a hydrophobic, protease-resistant core.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8UN1
 
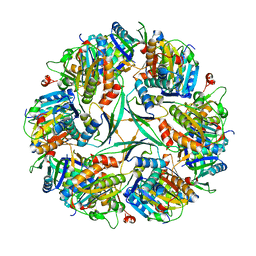 | | T33-ml23 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml23-redesigned-CutA-fold, T33-ml23-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
8UMR
 
 | | T33-ml35 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml35-redesigned-CutA-fold, T33-ml35-redesigned-TPR-domain-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
6V8R
 
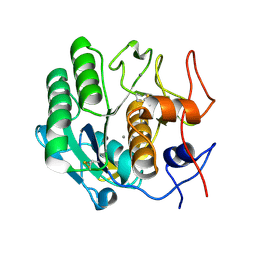 | | Proteinase K Determined by MicroED Phased by ARCIMBOLDO_SHREDDER | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Richards, L.S, Martynowycz, M.W, Sawaya, M.R, Millan, C. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.6 Å) | | Cite: | Fragment-based determination of a proteinase K structure from MicroED data using ARCIMBOLDO_SHREDDER
Acta Crystallogr.,Sect.D, 76, 2020
|
|
6UCX
 
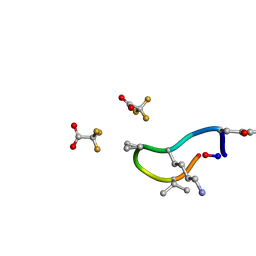 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDZ
 
 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UD9
 
 | | S2 symmetric peptide design number 2, Morticia | | Descriptor: | S2-2, Morticia | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UFR
 
 | | Structure of recombinantly assembled E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Li, B, Jiang, L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8V9O
 
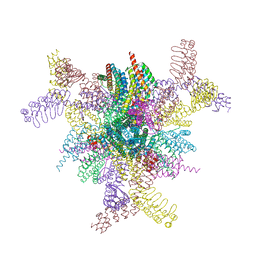 | | Imaging scaffold engineered to bind the therapeutic protein target BARD1 | | Descriptor: | CALCIUM ION, Tetrahedral Nanocage Cage Component Fused to Anti-BARD1 Darpin, Tetrahedral Nanocage Cage, ... | | Authors: | Agdanowski, M.P, Castells-Graells, R, Sawaya, M.R, Yeates, T.O, Arbing, M.A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | X-ray crystal structure of a designed rigidified imaging scaffold in the ligand-free conformation.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
6VW2
 
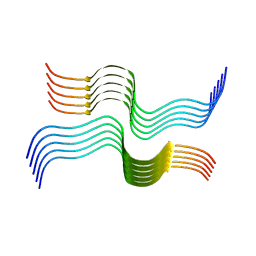 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils | | Descriptor: | Islet amyloid polypeptide (SUMO-tagged) | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2020-02-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure and inhibitor design of human IAPP (amylin) fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8UI2
 
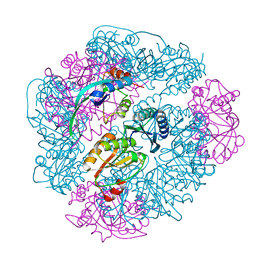 | | T33-ml28 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml28-redesigned-CutA-fold, T33-ml28-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-09 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
