3HPM
 
 | |
3HPK
 
 | |
1TIO
 
 | | HIGH PACKING DENSITY FORM OF BOVINE BETA-TRYPSIN IN CYCLOHEXANE | | Descriptor: | BENZAMIDINE, CALCIUM ION, CYCLOHEXANE, ... | | Authors: | Huang, Q, Zhu, G, Wang, Z, Tang, Q. | | Deposit date: | 1998-09-17 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray studies on two forms of bovine beta-trypsin crystals in neat cyclohexane.
Biochim.Biophys.Acta, 1429, 1998
|
|
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
6DK9
 
 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
6DKA
 
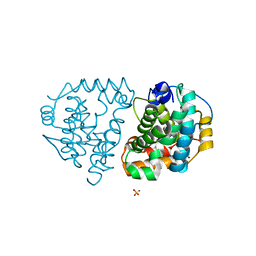 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | CYANAMIDE, DNA damage-inducible protein, SULFATE ION, ... | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
6DKD
 
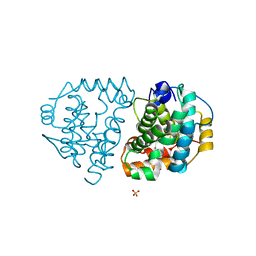 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
6DKC
 
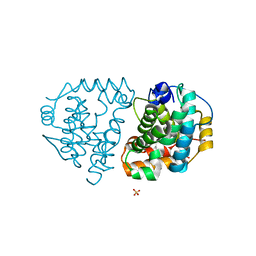 | | Yeast Ddi2 Cyanamide Hydratase, T157V mutant, apo structure | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
5FEQ
 
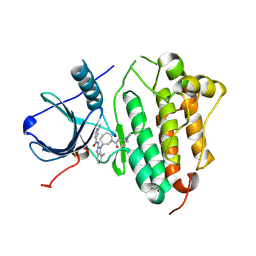 | | EGFR KINASE DOMAIN IN COMPLEX WITH A COVALENT AMINOBENZIMIDAZOLE | | Descriptor: | Epidermal growth factor receptor, ~{N}-[1-[(3~{R})-1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl]-7-methyl-benzimidazol-2-yl]-2-methyl-pyridine-4-carboxamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-17 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
1PYM
 
 | |
3LCT
 
 | |
3LCS
 
 | |
3L9P
 
 | |
3O17
 
 | | Crystal Structure of JNK1-alpha1 isoform | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, JIP1, 10MER PEPTIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2010-07-20 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of JNK1-alpha1 isoform
TO BE PUBLISHED
|
|
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8JDA
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | Descriptor: | Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
3V5Q
 
 | | Discovery of a selective TRK Inhibitor with efficacy in rodent cancer tumor models | | Descriptor: | 1-(3-{[(3Z)-2-oxo-3-(1H-pyrrol-2-ylmethylidene)-2,3-dihydro-1H-indol-6-yl]amino}phenyl)-3-[3-(trifluoromethyl)phenyl]urea, CHLORIDE ION, NT-3 growth factor receptor | | Authors: | Kreusch, A. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Discovery of GNF-5837, a Selective TRK Inhibitor with Efficacy in Rodent Cancer Tumor Models.
ACS Med Chem Lett, 3, 2012
|
|
4OUM
 
 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CITRATE ANION, Caprin-2, ISOPROPYL ALCOHOL, ... | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
4OUL
 
 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2, GLYCEROL | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
4OUS
 
 | | Crystal structure of zebrafish Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2 | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
4YMJ
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-[6-(benzylamino)imidazo[1,2-b]pyridazin-3-yl]benzonitrile, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YPS
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-{6-[(3R)-3-(3-fluorophenyl)morpholin-4-yl]imidazo[1,2-b]pyridazin-3-yl}benzonitrile, High affinity nerve growth factor receptor, SULFATE ION | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-13 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1012 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YNE
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 6-[(2R)-2-(3-fluorophenyl)pyrrolidin-1-yl]-3-(pyridin-2-yl)imidazo[1,2-b]pyridazine, GLYCEROL, High affinity nerve growth factor receptor, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0229 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
7WEB
 
 | |
7WEE
 
 | |
