3HJV
 
 | | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Malonyl Coa-acyl carrier protein transacylase, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Anderson, S, Skarina, T, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3FF1
 
 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3HZN
 
 | | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase | | Descriptor: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase
TO BE PUBLISHED
|
|
3IHS
 
 | | Crystal Structure of a Phosphocarrier Protein HPr from Bacillus anthracis str. Ames | | Descriptor: | Phosphocarrier protein HPr | | Authors: | Brunzelle, J.S, Anderson, S.M, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Phosphocarrier Protein HPr from Bacillus anthracis str. Ames
To be Published
|
|
3IG4
 
 | | Structure of a putative aminopeptidase P from Bacillus anthracis | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Xaa-pro aminopeptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a putative aminopeptidase P from Bacillus anthracis
To be Published
|
|
1NSL
 
 | | Crystal structure of Probable acetyltransferase | | Descriptor: | CHLORIDE ION, Probable acetyltransferase | | Authors: | Brunzelle, J.S, Korolev, S.V, Wu, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YdaF protein: A putative ribosomal N-acetyltransferase
Proteins, 57, 2004
|
|
1Q8I
 
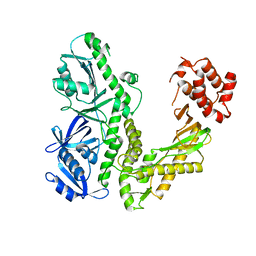 | | Crystal structure of ESCHERICHIA coli DNA Polymerase II | | Descriptor: | DNA polymerase II | | Authors: | Brunzelle, J.S, Muchmore, C.R.A, Mashhoon, N, Blair-Johnson, M, Shuvalova, L, Goodman, M.F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-21 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Escherichia Coli DNA Polymerase II
To be Published
|
|
1SFS
 
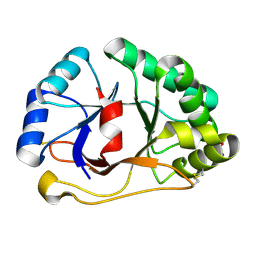 | | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein | | Descriptor: | Hypothetical protein, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein
To be Published
|
|
1RXQ
 
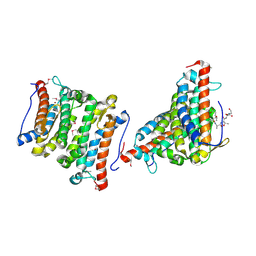 | | YfiT from Bacillus subtilis is a probable metal-dependent hydrolase with an unusual four-helix bundle topology | | Descriptor: | ALANINE, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Rajan, S.S, Yang, X, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-18 | | Release date: | 2004-07-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | YfiT from Bacillus subtilis Is a Probable Metal-Dependent Hydrolase with an Unusual Four-Helix Bundle Topology
Biochemistry, 43, 2004
|
|
1Q7R
 
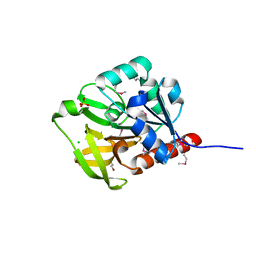 | |
1SF9
 
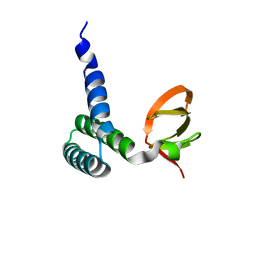 | | Crystal Structure of Bacillus subtilis YfhH Protein : Putative Transcriptional Regulator | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, yfhH hypothetical protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kim, D.E, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-19 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of Bacillus Subtilis YfhH hypothetical protein
To be Published
|
|
1SU1
 
 | | Structural and biochemical characterization of Yfce, a phosphoesterase from E. coli | | Descriptor: | Hypothetical protein yfcE, SULFATE ION, ZINC ION | | Authors: | Miller, D.J, Shuvalova, L, Evdokimova, E, Savchenko, A, Yakunin, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene.
Protein Sci., 16, 2007
|
|
1DUG
 
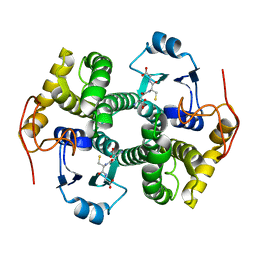 | | STRUCTURE OF THE FIBRINOGEN G CHAIN INTEGRIN BINDING AND FACTOR XIIIA CROSSLINKING SITES OBTAINED THROUGH CARRIER PROTEIN DRIVEN CRYSTALLIZATION | | Descriptor: | GLUTATHIONE, chimera of GLUTATHIONE S-TRANSFERASE-synthetic LINKEr-C-TERMINAL FIBRINOGEN GAMMA CHAIN | | Authors: | Ware, S, Donahue, J.P, Hawiger, J, Anderson, W.F. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the fibrinogen gamma-chain integrin binding and factor XIIIa cross-linking sites obtained through carrier protein driven crystallization.
Protein Sci., 8, 1999
|
|
1EXC
 
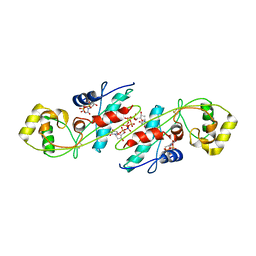 | | CRYSTAL STRUCTURE OF B. SUBTILIS MAF PROTEIN COMPLEXED WITH D-(UTP) | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, PROTEIN MAF, SODIUM ION | | Authors: | Minasov, G, Teplova, M, Stewart, G.C, Koonin, E.V, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EX2
 
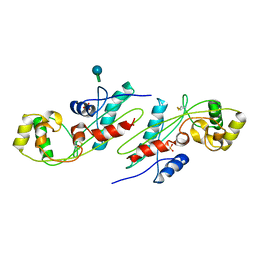 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS MAF PROTEIN | | Descriptor: | PHOSPHATE ION, PROTEIN MAF, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Minasov, G, Teplova, M, Stewart, G.C, Koonin, E.V, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5HI6
 
 | | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form
To Be Published
|
|
5HD6
 
 | | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL | | Authors: | Chang, C, Maltseva, N, Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A
To Be Published
|
|
5HZL
 
 | |
5HJ5
 
 | | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, ACETIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE -6-PHOSPHATE
To Be Published
|
|
5HX7
 
 | | Metal ABC transporter from Listeria monocytogenes | | Descriptor: | Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-29 | | Release date: | 2016-02-17 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes
to be published
|
|
5HVN
 
 | | 3.0 Angstrom Crystal Structure of 3-dehydroquinate Synthase (AroB) from Francisella tularensis in Complex with NAD. | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Zhou, M, Grimshaw, S, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3.0 Angstrom Crystal Structure of 3-dehydroquinate Synthase (AroB) from Francisella tularensis in Complex with NAD.
To Be Published
|
|
5HXM
 
 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with panose | | Descriptor: | Alpha-xylosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Winsor, J, Grimshaw, S, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-31 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5HOP
 
 | | 1.65 Angstrom resolution crystal structure of lmo0182 (residues 1-245) from Listeria monocytogenes EGD-e | | Descriptor: | ACETATE ION, Lmo0182 protein | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5HSF
 
 | | 1.52 Angstrom Crystal Structure of Fc fragment of Human IgG1. | | Descriptor: | Ig gamma-1 chain C region, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Flores, K, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-25 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | 1.52 Angstrom Crystal Structure of Fc fragment of Human IgG1.
To Be Published
|
|
5I0E
 
 | | Cycloalternan-degrading enzyme from Trueperella pyogenes in complex with isomaltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glycoside hydrolase family 31, ... | | Authors: | Light, S.H, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-03 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
