3IAU
 
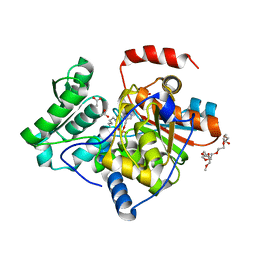 | | The structure of the processed form of threonine deaminase isoform 2 from Solanum lycopersicum | | Descriptor: | ACETATE ION, POLYETHYLENE GLYCOL (N=34), SULFATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Adaptive evolution of threonine deaminase in plant defense against insect herbivores.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8GCT
 
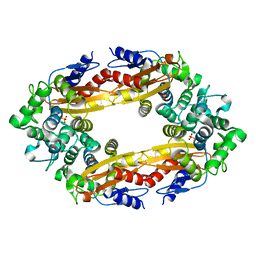 | |
8GCS
 
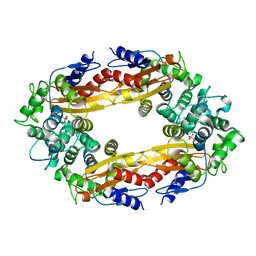 | |
8GCV
 
 | |
8GCX
 
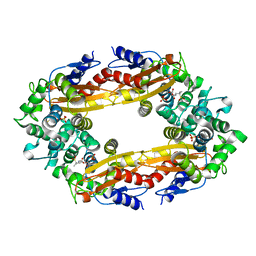 | |
6OMR
 
 | | Crystal structure of PtmU3 complexed with PTN substrate | | Descriptor: | ACETATE ION, MANGANESE (II) ION, PtmU3, ... | | Authors: | Liu, Y.C, Dong, L.B, Shen, B. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Characterization and Crystal Structure of a Nonheme Diiron Monooxygenase Involved in Platensimycin and Platencin Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
6OMQ
 
 | | Crystal structure of PtmU3 complexed with PTM substrate | | Descriptor: | ACETATE ION, MANGANESE (II) ION, PtmU3, ... | | Authors: | Liu, Y.C, Dong, L.B, Shen, B. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterization and Crystal Structure of a Nonheme Diiron Monooxygenase Involved in Platensimycin and Platencin Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
1BAP
 
 | |
2IFU
 
 | | Crystal Structure of a Gamma-SNAP from Danio rerio | | Descriptor: | SULFATE ION, gamma-snap | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family.
Proteins, 70, 2008
|
|
2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | Descriptor: | COENZYME A, Protein At1g77540 | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
1APB
 
 | |
6OMP
 
 | | Crystal structure of apo PtmU3 | | Descriptor: | ACETATE ION, MANGANESE (II) ION, PtmU3 | | Authors: | Liu, Y.C, Dong, L.B, Shen, B. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and Crystal Structure of a Nonheme Diiron Monooxygenase Involved in Platensimycin and Platencin Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
2ATF
 
 | | X-RAY STRUCTURE OF cysteine dioxygenase type I FROM MUS MUSCULUS MM.241056 | | Descriptor: | 1,2-ETHANEDIOL, Cysteine dioxygenase type I, NICKEL (II) ION | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of mouse cysteine dioxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1ZTP
 
 | | X-ray structure of gene product from homo sapiens Hs.433573 | | Descriptor: | Basophilic leukemia expressed protein BLES03 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure at 2.5 A resolution of human basophilic leukemia-expressed protein BLES03.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2BEI
 
 | | X-ray structure of thialysine n-acetyltransferase (SSAT2) from homo sapiens | | Descriptor: | ACETYL COENZYME *A, Diamine acetyltransferase 2 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Crystal structure of Homo sapiens thialysine Nepsilon-acetyltransferase (HsSSAT2) in complex with acetyl coenzyme A.
Proteins, 64, 2006
|
|
3BVO
 
 | | Crystal structure of human co-chaperone protein HscB | | Descriptor: | Co-chaperone protein HscB, mitochondrial precursor, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-01-07 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain.
J.Biol.Chem., 283, 2008
|
|
2BDU
 
 | | X-Ray Structure of a Cytosolic 5'-Nucleotidase III from Mus Musculus MM.158936 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2LUZ
 
 | | Solution NMR Structure of CalU16 from Micromonospora echinospora, Northeast Structural Genomics Consortium (NESG) Target MiR12 | | Descriptor: | CalU16 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Pederson, K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Wrobel, R.L, Bingman, C.A, Singh, S, Thorson, J.S, Prestegard, J.H, Montelione, G.T, Phillips Jr, G.N, Kennedy, M.A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
2APJ
 
 | | X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution | | Descriptor: | Putative Esterase | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-16 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure at 1.6 Angstroms resolution of the protein product of the At4g34215 gene from Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A13
 
 | | X-ray structure of protein from Arabidopsis thaliana AT1G79260 | | Descriptor: | At1g79260 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-17 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1.
Proteins, 78, 2010
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
8EBI
 
 | |
8EC4
 
 | |
8ECF
 
 | |
8EBR
 
 | |
