7Q0C
 
 | | Mimic carbonic anhydrase IX in complex with Methyl 2-chloro-4-(cyclohexylsulfanyl)-5-sulfamoylbenzoate | | Descriptor: | ACETATE ION, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
7OZ3
 
 | | S. agalactiae BusR in complex with its busA-promotor DNA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GntR family transcriptional regulator, pBusA_for, ... | | Authors: | Bandera, A.M, Witte, G. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res., 49, 2021
|
|
8RBP
 
 | | Crystal structure of chimeric human carbonic anhydrase IX with 4-chloro-2-(cyclohexylsulfanyl)-N-(2-hydroxyethyl)-5-sulfamoylbenzamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-chloranyl-2-cyclohexylsulfanyl-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 2, ... | | Authors: | Manakova, E.N, Smirnov, A, Paketuryte, V, Grazulis, S. | | Deposit date: | 2023-12-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | From X-ray crystallographic structure to intrinsic thermodynamics of protein-ligand binding using carbonic anhydrase isozymes as a model system.
Iucrj, 11, 2024
|
|
8RJ2
 
 | | Crystal structure of carbonic anhydrase II with N-butyl-4-chloro-2-(cyclohexylsulfanyl)-5-sulfamoylbenzamide | | Descriptor: | BICINE, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E.N, Grazulis, S. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | From X-ray crystallographic structure to intrinsic thermodynamics of protein-ligand binding using carbonic anhydrase isozymes as a model system.
Iucrj, 11, 2024
|
|
8P9E
 
 | | Crystal structure of wild type p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 F11 | | Descriptor: | Darpin 1810 F11, GLYCEROL, Isoform 2 of Tumor protein 63, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
2YNM
 
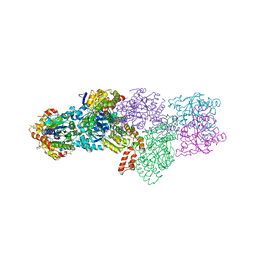 | | Structure of the ADPxAlF3-Stabilized Transition State of the Nitrogenase-like Dark-Operative Protochlorophyllide Oxidoreductase Complex from Prochlorococcus marinus with Its Substrate Protochlorophyllide a | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Krausze, J, Lange, C, Heinz, D.W, Moser, J. | | Deposit date: | 2012-10-16 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Adp-Aluminium Fluoride-Stabilized Protochlorophyllide Oxidoreductase Complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
9HK1
 
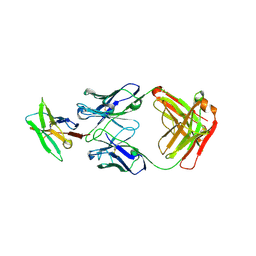 | | PD1 signaling receptor bound to FAB Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody FAB heavy chain, Antibody FAB light chain, ... | | Authors: | Bjorkelid, C, Paluch, C, Robertson, N.J. | | Deposit date: | 2024-12-02 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Antibody agonists trigger immune receptor signaling through local exclusion of receptor-type protein tyrosine phosphatases.
Immunity, 57, 2024
|
|
8EB1
 
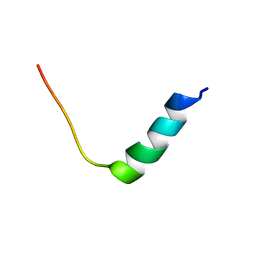 | | Chim2 - Intragenic antimicrobial peptide | | Descriptor: | Unconventional myosin-Ih, Transcription activator BRG1 intragenic antimicrobial chimeric peptide | | Authors: | de Freitas, T.V, Oliveira, A.L, Santos, M.A, Brand, G.D. | | Deposit date: | 2022-08-30 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Release of immunomodulatory peptides at bacterial membrane interfaces as a novel strategy to fight microorganisms.
J.Biol.Chem., 299, 2023
|
|
5L92
 
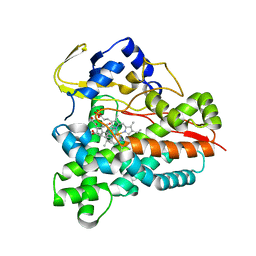 | |
5L91
 
 | |
5L90
 
 | |
6Y4D
 
 | | Crystal structure of a short-chain dehydrogenase/reductase (SDR) from Zephyranthes treatiae in complex with NADP+ | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain dehydrogenase/reductase (SDR) | | Authors: | Sautner, V, Steimle, S, Roth, S, Mueller, M, Tittmann, K. | | Deposit date: | 2020-02-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crossing the Border: From Keto- to Imine Reduction in Short-Chain Dehydrogenases/Reductases.
Chembiochem, 21, 2020
|
|
6Q57
 
 | |
5L94
 
 | |
2IAS
 
 | | Crystal structure of squid ganglion DFPase W244F mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
2IAR
 
 | | Crystal structure of squid ganglion DFPase W244H mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
2IAQ
 
 | | Crystal structure of squid ganglion DFPase S271A mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
2IAW
 
 | | Crystal structure of squid ganglion DFPase N175D mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Katsemi, V, Luecke, C, Koepke, J, Loehr, F, Maurer, S, Fritzsch, G, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mutational and structural studies of the diisopropylfluorophosphatase from Loligo vulgaris shed new light on the catalytic mechanism of the enzyme
Biochemistry, 44, 2005
|
|
4R4S
 
 | | Crystal structure of chimeric beta-lactamase cTEM-19m at 1.1 angstrom resolution | | Descriptor: | Beta-lactamase TEM,Beta-lactamase PSE-4, CHLORIDE ION, MAGNESIUM ION | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-08-19 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Structural Dynamics of Engineered beta-Lactamases Vary Broadly on Three Timescales yet Sustain Native Function.
Sci Rep, 9, 2019
|
|
8RAR
 
 | | Crystal structure of chimeric human carbonic anhydrase IX with N-butyl-4-chloro-2-(cyclohexylsulfanyl)-5-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Manakova, E.N, Smirnov, A, Paketuryte, V, Grazulis, S. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | From X-ray crystallographic structure to intrinsic thermodynamics of protein-ligand binding using carbonic anhydrase isozymes as a model system.
Iucrj, 11, 2024
|
|
4MEZ
 
 | | Crystal structure of M68L/M69T double mutant TEM-1 | | Descriptor: | Beta-lactamase TEM, CHLORIDE ION, GLYCEROL, ... | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2013-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | The Structural Dynamics of Engineered beta-Lactamases Vary Broadly on Three Timescales yet Sustain Native Function.
Sci Rep, 9, 2019
|
|
4R4R
 
 | | Crystal structure of chimeric beta-lactamase cTEM-19m at 1.2 angstrom resolution | | Descriptor: | Beta-lactamase TEM,Beta-lactamase PSE-4, CHLORIDE ION, MAGNESIUM ION | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-08-19 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structural Dynamics of Engineered beta-Lactamases Vary Broadly on Three Timescales yet Sustain Native Function.
Sci Rep, 9, 2019
|
|
2L22
 
 | | Mupirocin didomain ACP | | Descriptor: | Mupirocin didomain Acyl Carrier Protein | | Authors: | Dong, X, Williams, C, Crump, M.P, Wattana-amorn, P. | | Deposit date: | 2010-08-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A conserved motif flags acyl carrier proteins for beta-branching in polyketide synthesis.
Nat.Chem.Biol., 9, 2013
|
|
4NCH
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 L802W mutation | | Descriptor: | DNA double-strand break repair Rad50 ATPase, SULFATE ION | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4NCK
 
 | | Crystal Structure of Pyrococcus furiosis Rad50 R797G mutation | | Descriptor: | CHLORIDE ION, DNA double-strand break repair Rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
