7WR7
 
 | | Structure of Inhibited-EP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Enteropeptidase catalytic light chain, ... | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7V55
 
 | |
7V53
 
 | |
6A0A
 
 | |
6A0C
 
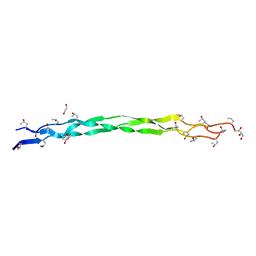 | | Structure of a triple-helix region of human collagen type III | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, collagen type III peptide | | Authors: | Yang, X, Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Characterization by high-resolution crystal structure analysis of a triple-helix region of human collagen type III with potent cell adhesion activity.
Biochem. Biophys. Res. Commun., 508, 2019
|
|
6CHW
 
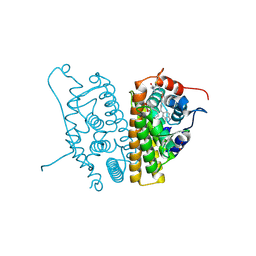 | | Estrogen Receptor Alpha Y537S covalently bound to antagonist H3B-5942. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
6CHZ
 
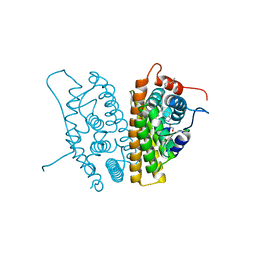 | | Estrogen Receptor Alpha Y537S bound to antagonist H3B-9224. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, Estrogen receptor | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
8VI1
 
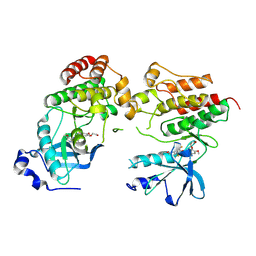 | | Crystal structure of c-Met-D1228N in complex with KIN-7615 | | Descriptor: | Hepatocyte growth factor receptor, N-(3,5-difluoro-4-{[6-(2-hydroxyethoxy)-7-methoxyquinolin-4-yl]oxy}phenyl)-4-methoxypyridine-3-carboxamide | | Authors: | Ouyang, X, Kania, R, Cox, J. | | Deposit date: | 2024-01-02 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Discovery of KIN-8741, a Highly Selective Type IIb c-Met Kinase Inhibitor with Broad Mutation Coverage and Quality Drug-Like Properties for the Treatment of Cancer.
J.Med.Chem., 68, 2025
|
|
4U7Z
 
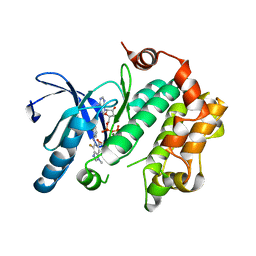 | | Mitogen-Activated Protein Kinase Kinase (MEK1) bound to G805 | | Descriptor: | 5-[(4-bromo-2-chlorophenyl)amino]-4-fluoro-N-(2-hydroxyethoxy)-1-methyl-1H-benzimidazole-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Robarge, K.D, Ultsch, M.H, Wiesmann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U80
 
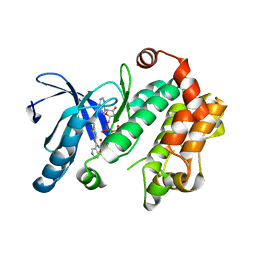 | | MEK 1 kinase bound to G799 | | Descriptor: | 3-[(4-cyclopropyl-2-fluorophenyl)amino]-N-(2-hydroxyethoxy)furo[3,2-c]pyridine-2-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ultsch, M.H, Robarge, K.D, Weismann, C. | | Deposit date: | 2014-07-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
9L0Q
 
 | |
7F3B
 
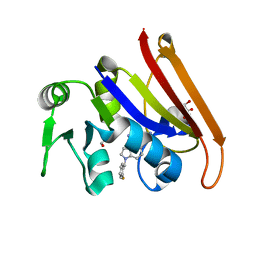 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
9LJ3
 
 | | Crystal structure of P25 | | Descriptor: | 3-[[4-chloranyl-2-[(2-hydroxyethylamino)methyl]-5-[[2-methyl-3-[3-[2-[2-[(3~{R})-3-oxidanylpyrrolidin-1-yl]ethoxy]ethoxy]phenyl]phenyl]methoxy]phenoxy]methyl]benzenecarbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Yang, P, Chen, M.R, Xiao, Y.B. | | Deposit date: | 2025-01-14 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery and Crystallography Study of Novel Resorcinol Dibenzyl Ether-Based PD-1/PD-L1 Inhibitors with Improved Drug-like and Pharmacokinetic Properties for Cancer Treatment.
J.Med.Chem., 68, 2025
|
|
3DJ3
 
 | |
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-01-26 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
6VNS
 
 | | Crystal structure of TYK2 kinase with compound 13 | | Descriptor: | (1R,2R)-2-cyano-N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6VNY
 
 | | Crystal structure of TYK2 kinase with compound 10 | | Descriptor: | N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropanecarboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6VNV
 
 | | Crystal structure of TYK2 kinase with compound 14 | | Descriptor: | (1S,2S)-2-cyano-N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6VNX
 
 | | Crystal structure of TYK2 kinase with compound 19 | | Descriptor: | (1S)-2,2-difluoro-N-[(1S,5R,6R)-3-{5-fluoro-2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-6-methyl-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6W8L
 
 | | Crystal structure of JAK1 kinase with compound 10 | | Descriptor: | N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropanecarboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
3SJ1
 
 | | PpcA M58D mutant | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CYTOCHROME C7, HEME C, ... | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pitfalls in the interpretation of structural changes in mutant proteins from crystal structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3SJ0
 
 | | PpcA mutant M58S | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CYTOCHROME C7, HEME C, ... | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pitfalls in the interpretation of structural changes in mutant proteins from crystal structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3SJ4
 
 | | PpcA mutant M58K | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CYTOCHROME C7, HEME C, ... | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pitfalls in the interpretation of structural changes in mutant proteins from crystal structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3SEL
 
 | | PpcA M58N mutant | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CYTOCHROME C7, HEME C, ... | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pitfalls in the interpretation of structural changes in mutant proteins from crystal structures.
J.Struct.Funct.Genom., 13, 2012
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | Descriptor: | CidA_I gamma/2 protein, CidB_I b/2 protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
