4IQF
 
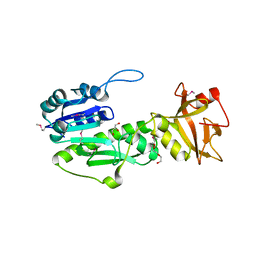 | | Crystal Structure of Methyionyl-tRNA Formyltransferase from Bacillus anthracis | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, GLYCEROL, Methionyl-tRNA formyltransferase, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Crystal Structure of Methyionyl-tRNA Formyltransferase from Bacillus anthracis
To be Published
|
|
4IR0
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
4JB7
 
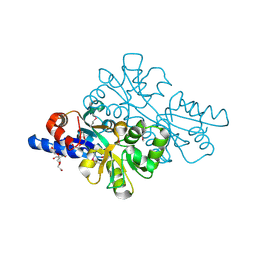 | | 1.42 Angstrom resolution crystal structure of accessory colonization factor AcfC (acfC) in complex with D-aspartic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Accessory colonization factor AcfC, D-MALATE, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Dubrovska, I, Winsor, J, Minasov, G, Shuvalova, L, Filippova, E.V, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42 Angstrom resolution crystal structure of accessory colonization factor AcfC (acfC) in complex with D-aspartic acid
To be Published
|
|
3M8A
 
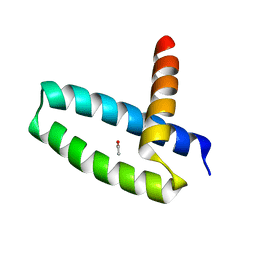 | | Crystal Structure of Swine Flu Virus NS1 N-Terminal RNA Binding Domain from H1N1 Influenza A/California/07/2009 | | Descriptor: | ACETATE ION, MALONATE ION, Nonstructural protein 1, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Swine Flu Virus NS1 N-Terminal RNA Binding Domain from H1N1 Influenza A/California/07/2009
To be Published
|
|
3HMQ
 
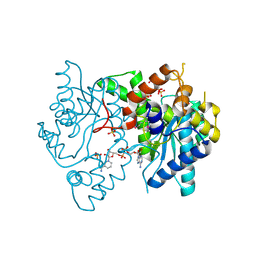 | | 1.9 Angstrom resolution crystal structure of a NAD synthetase (nadE) from Salmonella typhimurium LT2 in complex with NAD(+) | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of a NAD synthetase (nadE) from Salmonella typhimurium LT2 in complex with NAD(+)
To be Published
|
|
3N3W
 
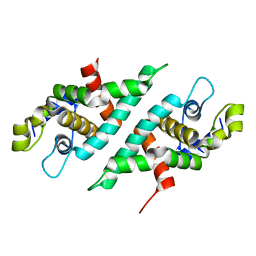 | | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni | | Descriptor: | Ribonuclease III | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni
TO BE PUBLISHED
|
|
3G48
 
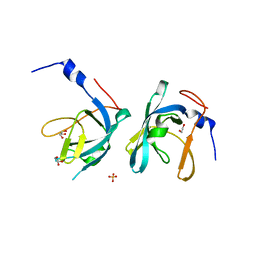 | | Crystal structure of chaperone CsaA form Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, Chaperone CsaA, GLYCEROL, ... | | Authors: | Nocek, B, Zhou, M, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-03 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of chaperone CsaA form Bacillus anthracis str. Ames
TO BE PUBLISHED
|
|
3M5R
 
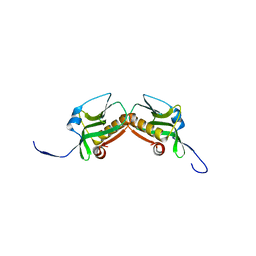 | |
3MJD
 
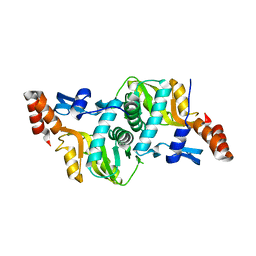 | | 1.9 Angstrom Crystal Structure of Orotate Phosphoribosyltransferase (pyrE) Francisella tularensis. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Orotate phosphoribosyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-12 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Orotate Phosphoribosyltransferase (pyrE) Francisella tularensis.
TO BE PUBLISHED
|
|
3IMI
 
 | | 2.01 Angstrom resolution crystal structure of a HIT family protein from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | HIT family protein, SULFATE ION, ZINC ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Gordon, E, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2.01 Angstrom resolution crystal structure of a HIT family protein from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3IJR
 
 | | 2.05 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD+ | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase, ... | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Gordon, E, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD+
To be Published
|
|
3HJV
 
 | | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Malonyl Coa-acyl carrier protein transacylase, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Anderson, S, Skarina, T, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
4KWU
 
 | | 1.9 Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e in complex with alpha-D-glucose, beta-D-glucose, magnesium and calcium | | Descriptor: | CALCIUM ION, Lmo2446 protein, MAGNESIUM ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-05-24 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e in complex with alpha-D-glucose, beta-D-glucose, magnesium and calcium
To be Published
|
|
3MGA
 
 | | 2.4 Angstrom Crystal Structure of Ferric Enterobactin Esterase (fes) from Salmonella typhimurium | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Enterochelin esterase, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of Ferric Enterobactin Esterase (fes) from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3NAV
 
 | | Crystal structure of an alpha subunit of tryptophan synthase from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, Tryptophan synthase alpha chain | | Authors: | Nocek, B, Makowska-Grzyska, M, Kwon, K, Anderson, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-06-02 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha subunit of tryptophan synthase from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
4GUC
 
 | | 1.4 Angstrom resolution crystal structure of uncharacterized protein BA_2500 from Bacillus anthracis str. Ames | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Protein BA_2500, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 Angstrom resolution crystal structure of uncharacterized protein BA_2500 from Bacillus anthracis str. Ames
To be Published
|
|
4H3D
 
 | | 1.95 Angstrom Crystal Structure of of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Modified Comenic Acid. | | Descriptor: | 3-dehydroquinate dehydratase, 5-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid, ACETATE ION, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Duban, M.-E, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Modified Comenic Acid.
TO BE PUBLISHED
|
|
4JD1
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | FOSFOMYCIN, Metallothiol transferase FosB 2, TRIETHYLENE GLYCOL, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Sharma, S.V, Hamilton, C.J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
3K28
 
 | | Crystal Structure of a glutamate-1-semialdehyde aminotransferase from Bacillus anthracis with bound Pyridoxal 5'Phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glutamate-1-semialdehyde 2,1-aminomutase 2, ... | | Authors: | Sharma, S.S, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-29 | | Release date: | 2010-01-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a glutamate-1-semialdehyde aminotransferase from Bacillus anthracis with bound Pyridoxal 5'Phosphate
To be Published
|
|
4HV4
 
 | | 2.25 Angstrom resolution crystal structure of UDP-N-acetylmuramate--L-alanine ligase (murC) from Yersinia pestis CO92 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom resolution crystal structure of UDP-N-acetylmuramate--L-alanine ligase (murC) from Yersinia pestis CO92 in complex with AMP
To be Published
|
|
4IFA
 
 | | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Shatsman, S, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-14 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames
To be Published
|
|
4LES
 
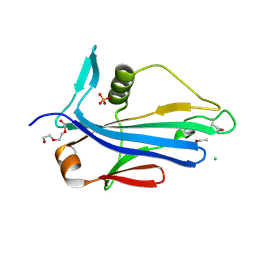 | | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, ETHANOL, PHOSPHATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Filippova, E, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
3H07
 
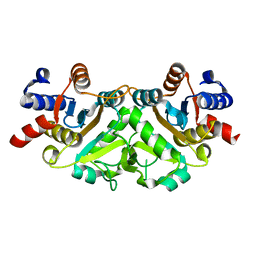 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Yersinia pestis CO92 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase | | Authors: | Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-08 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Yersinia pestis CO92
To be Published
|
|
4IJK
 
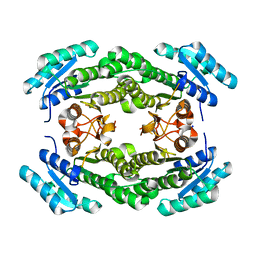 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase (FabG), SODIUM ION | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I.G, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695
To be Published
|
|
4IIN
 
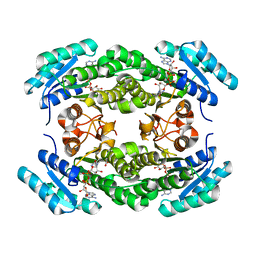 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+ | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase (FabG), ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+
To be Published
|
|
