2OB9
 
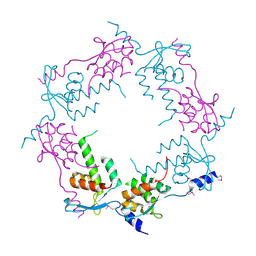 | | Structure of bacteriophage HK97 tail assembly chaperone | | Descriptor: | Tail assembly chaperone | | Authors: | McGrath, T.E, Tuite, A, Bona, D, Saridakis, V, Edwards, A.M, Maxwell, K, Chirgadze, N.Y. | | Deposit date: | 2006-12-18 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conserved spiral structure for highly diverged phage tail assembly chaperones.
J.Mol.Biol., 425, 2013
|
|
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | Descriptor: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
5FBM
 
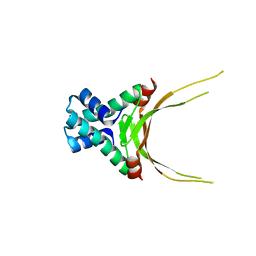 | | Crystal Structure of Histone Like Protein (HLP) from Streptococcus mutans Refined to 1.9 A Resolution | | Descriptor: | DNA-binding protein HU | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, O'Neil, P, Biswas, I. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of histone-like protein from Streptococcus mutans refined to 1.9 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
3CXN
 
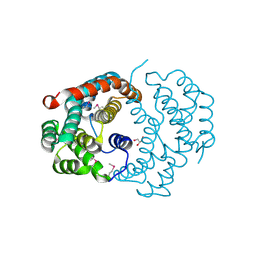 | | Structure of the Urease Accessory Protein UreF from Helicobacter pylori | | Descriptor: | GLYCEROL, Urease accessory protein ureF | | Authors: | Lam, R, Johns, K, Romanov, V, Dong, A, Wu-Brown, J, Guthrie, J, Dharamsi, A, Thambipillai, D, Mansoury, K, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2008-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a truncated urease accessory protein UreF from Helicobacter pylori.
Proteins, 78, 2010
|
|
3T3S
 
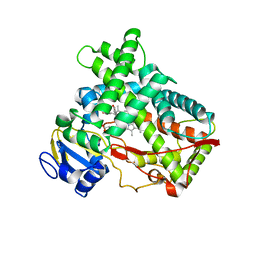 | | Human Cytochrome P450 2A13 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A13, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
3T3R
 
 | | Human Cytochrome P450 2A6 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
3T3Q
 
 | | Human Cytochrome P450 2A6 I208S/I300F/G301A/S369G in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
3T3Z
 
 | | Human Cytochrome P450 2E1 in complex with pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2E1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Meneely, K.M, DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
6DE6
 
 | |
3KOH
 
 | | Cytochrome P450 2E1 with omega-imidazolyl octanoic acid | | Descriptor: | 8-(1H-imidazol-1-yl)octanoic acid, Cytochrome P450 2E1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, Porubsky, P.R. | | Deposit date: | 2009-11-13 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Human cytochrome P450 2E1 structures with fatty acid analogs reveal a previously unobserved binding mode.
J.Biol.Chem., 285, 2010
|
|
3LC4
 
 | |
4DC7
 
 | | Crystal Structure of Myoglobin Exposed to Excessive SONICC Imaging Laser Dose. | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Becker, M, Mulichak, A.M, Kissick, D.J, Fischetti, R.F, Keefe, L.J, Simpson, G.J. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Towards protein-crystal centering using second-harmonic generation (SHG) microscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8DOR
 
 | |
8DOP
 
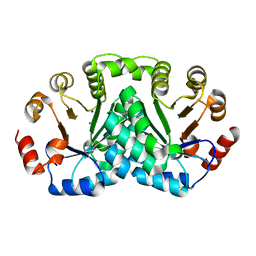 | |
8DOS
 
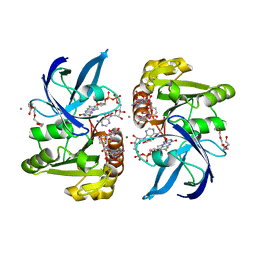 | |
8DQC
 
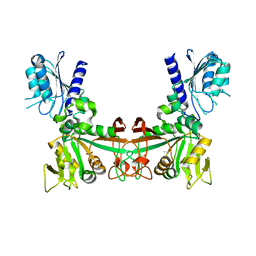 | |
8DOQ
 
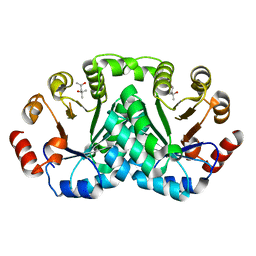 | |
8DQ9
 
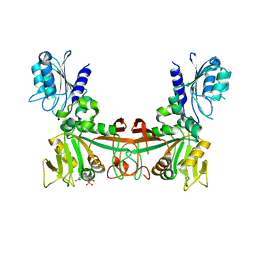 | |
8DQB
 
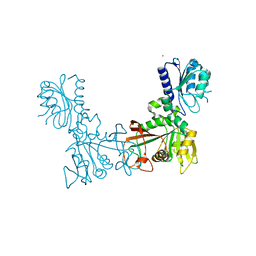 | |
8DUR
 
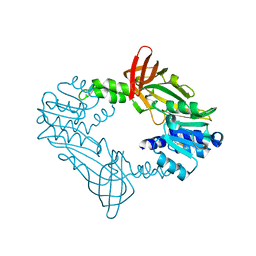 | |
6V02
 
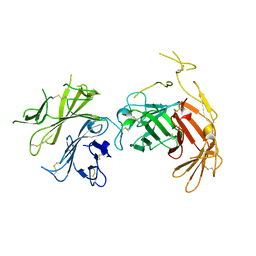 | | N-terminal 5 domains of CI-MPR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor | | Authors: | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol, 3, 2020
|
|
9B22
 
 | |
9B21
 
 | | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form) | | Descriptor: | ADP-ribose pyrophosphatase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-03-14 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form)
To be published
|
|
9B1Z
 
 | |
9AV6
 
 | |
