7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
8WZQ
 
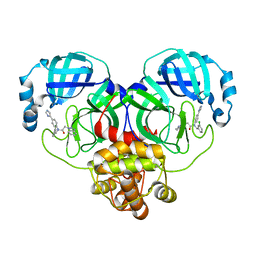 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with CCF0058981 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Jiang, H.H, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
8WZP
 
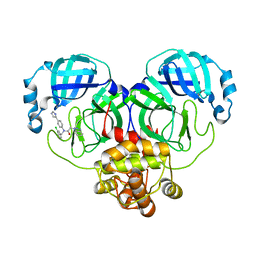 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with CCF0058981 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | Authors: | Jiang, H.H, Zou, X.F, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
8WUL
 
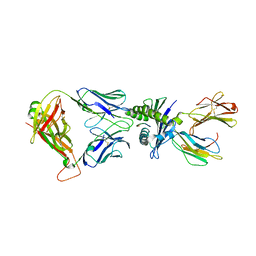 | | Crystal structure of affinity enhanced TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen, ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
8WTE
 
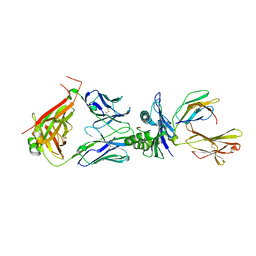 | | Crystal structure of TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen (Fragment), ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
5GNU
 
 | | the structure of mini-MFN1 apo | | Descriptor: | Mitofusin-1 | | Authors: | Yan, L, Yu, C, Ming, Z, Lou, Z, Rao, Z, Lou, J. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.113 Å) | | Cite: | BDLP-like folding of Mitofusin 1
To Be Published
|
|
8IG4
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with GC376 | | Descriptor: | Non-structural protein 11, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
9IKQ
 
 | |
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
9L0D
 
 | | Cryo-EM structure of the human MON1A/CCZ1/C18orf8 complex | | Descriptor: | Ras-related protein Rab-7a, Regulator of MON1-CCZ1 complex, Vacuolar fusion protein CCZ1 homolog B, ... | | Authors: | Tang, Y, Han, Y, Zhang, Y, Pan, L. | | Deposit date: | 2024-12-12 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanistic insights into the GEF activity of the human MON1A/CCZ1/C18orf8 complex.
Protein Cell, 2025
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
8SX3
 
 | | 10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10 | | Descriptor: | 10E8 Fab heavy chain, 10E8 light chain, 10E8-GT10.2 immunogen, ... | | Authors: | Huang, J, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Vaccination induces broadly neutralizing antibody precursors to HIV gp41.
Nat.Immunol., 25, 2024
|
|
6XQI
 
 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | Descriptor: | ASN-PRO-LEU-GLU-PHE-LEU, Protein Vpr, UV excision repair protein RAD23 homolog A, ... | | Authors: | Calero, G.C, Wu, Y, Weiss, S.C. | | Deposit date: | 2020-07-09 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
6XQJ
 
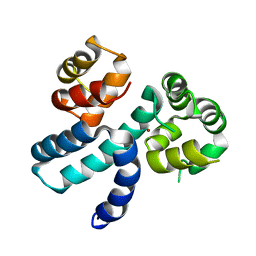 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | Descriptor: | Protein Vpr,UV excision repair protein RAD23 homolog A, ZINC ION | | Authors: | Byeon, I.-J.L, Calero, G, Wu, Y, Byeon, C.H, Gronenborn, A.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
6K0Y
 
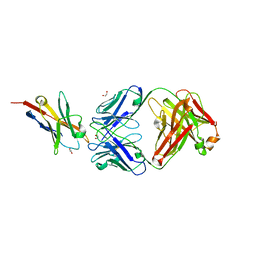 | | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Liu, J.X, Wang, G.Q. | | Deposit date: | 2019-05-08 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor.
Sci Rep, 9, 2019
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
8WLI
 
 | |
8WL2
 
 | | Cryo-EM structure of the membrane-anchored part of the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar P-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WK3
 
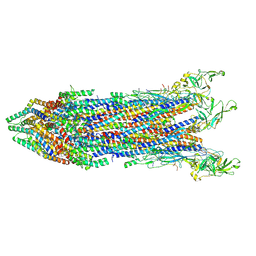 | | Cryo-EM structure of the proximal rod-export apparatus and FlgF within the motor-hook complex in the CW state | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-26 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WLE
 
 | | Cryo-EM structure of the LP ring within the flagellar motor-hook complex in the CCW state. | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WKQ
 
 | | Cryo-EM structure of the MS ring (C1) with export apparatus and proximal rod within the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WLH
 
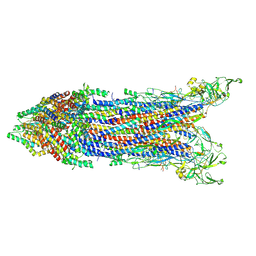 | | Cryo-EM structure of the proximal rod-export apparatus and FlgF within the motor-hook complex in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WJR
 
 | |
