6MB0
 
 | |
9DUG
 
 | |
9DR6
 
 | |
9DUF
 
 | |
9DU6
 
 | |
9DRF
 
 | |
9DRE
 
 | |
9DRD
 
 | |
9DU9
 
 | |
9DUA
 
 | |
9DT7
 
 | |
9DR8
 
 | |
9DRA
 
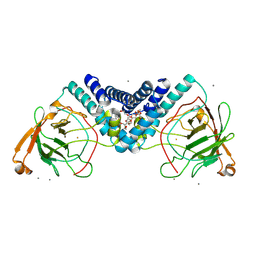 | | Crystal structure of Catechol 1,2-dioxygenase from Burkholderia multivorans (Iron and 4,5-dichloro-1,2-catechol bound) | | Descriptor: | (6R)-3,4-dichloro-6-hydroxycyclohex-3-en-1-one, CALCIUM ION, Catechol 1,2-dioxygenase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-09-25 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of Catechol 1,2-dioxygenase from Burkholderia multivorans (Iron and 4,5-dichloro-1,2-catechol bound)
To be published
|
|
9DTC
 
 | |
9DSZ
 
 | |
9DU8
 
 | |
9DT8
 
 | |
6MAI
 
 | |
6MAO
 
 | |
3JS5
 
 | |
7RHE
 
 | |
3KC6
 
 | |
3KHW
 
 | |
3KE1
 
 | |
4N67
 
 | |
