1AJA
 
 | |
4EWO
 
 | | Design and synthesis of potent hydroxyethylamine (hea) bace-1 inhibitors | | Descriptor: | Beta-secretase 1, N-[(2S,3R)-4-{[(4S)-2-(2,2-dimethylpropyl)-6,6-dimethyl-4,5,6,7-tetrahydro-2H-indazol-4-yl]amino}-3-hydroxy-1-phenylbutan-2-yl]acetamide | | Authors: | Borkakoti, N, Lindberg, J, Derbyshire, D. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of potent hydroxyethylamine (HEA) BACE-1 inhibitors carrying prime side 4,5,6,7-tetrahydrobenzazole and 4,5,6,7-tetrahydropyridinoazole templates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EXG
 
 | | Design and synthesis of potent hydroxyethylamine (hea) bace-1 inhibitors | | Descriptor: | Beta-secretase 1, N-[(2S,3R)-4-{[(4S)-6-(2,2-dimethylpropyl)-2,2-dimethyl-3,4-dihydro-2H-thieno[2,3-b]pyran-4-yl]amino}-3-hydroxy-1-phenylbutan-2-yl]acetamide | | Authors: | Borkakoti, N, Lindberg, J, Derbyshire, D. | | Deposit date: | 2012-04-30 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of potent hydroxyethylamine (HEA) BACE-1 inhibitors carrying prime side 4,5,6,7-tetrahydrobenzazole and 4,5,6,7-tetrahydropyridinoazole templates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6SG2
 
 | | FeFe Hydrogenase from Desulfovibrio desulfuricans in Hinact state | | Descriptor: | HydB, IRON/SULFUR CLUSTER, Periplasmic [Fe] hydrogenase large subunit, ... | | Authors: | Galle, L.M, Span, I. | | Deposit date: | 2019-08-02 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Caught in the H inact : Crystal Structure and Spectroscopy Reveal a Sulfur Bound to the Active Site of an O 2 -stable State of [FeFe] Hydrogenase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3TLU
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' oxidized mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TLW
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' oxidized mutant in a locally-closed conformation (LC2 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6SXI
 
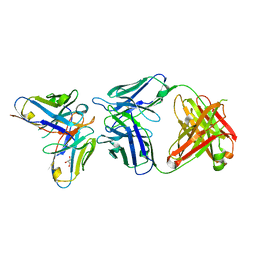 | | Antibody-anti-idiotype complex: AP33 Fab (hepatitis C virus E2 antibody) - B2.1A scFv (anti-idiotype) | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Taylor, G.L, Potter, J.A, Fadda, V, Patel, A.H, Owsianka, A.M, Cowtan, V.M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-10-07 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of a structural epitope mimic: an idiotypic approach to HCV vaccine design.
NPJ Vaccines, 6, 2021
|
|
3TLS
 
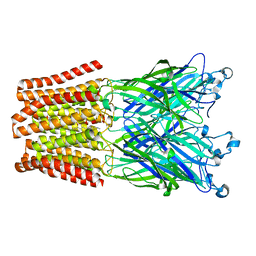 | | The GLIC pentameric Ligand-Gated Ion Channel E19'P mutant in a locally-closed conformation (LC2 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TLT
 
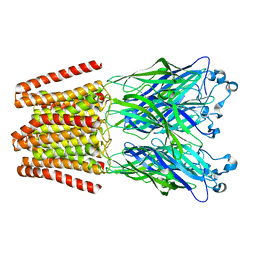 | | The GLIC pentameric Ligand-Gated Ion Channel H11'F mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TLV
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-22' oxidized mutant in a locally-closed conformation (LC3 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6G40
 
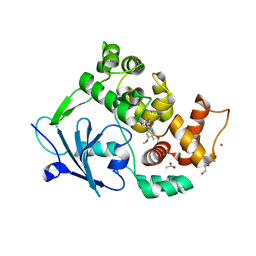 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH9525 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
1W87
 
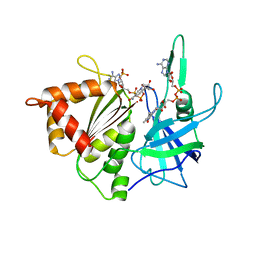 | |
3UU4
 
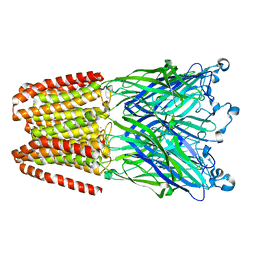 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in the crystal in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UUB
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UU3
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-20' oxidized mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UU5
 
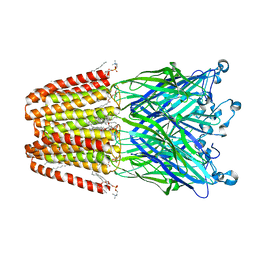 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-20' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UU8
 
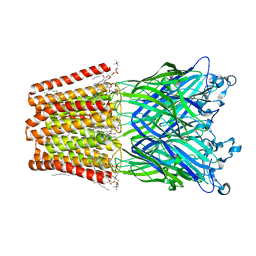 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UU6
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-22' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
8E5F
 
 | | Cryo-EM of P. calidifontis cytochrome filament | | Descriptor: | HEME C, c-type cytochrome | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Extracellular cytochrome nanowires appear to be ubiquitous in prokaryotes.
Cell, 186, 2023
|
|
8E5G
 
 | | Cryo-EM of A. veneficus cytochrome filament | | Descriptor: | HEME C, c-type cytochrome | | Authors: | Wang, F, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Extracellular cytochrome nanowires appear to be ubiquitous in prokaryotes.
Cell, 186, 2023
|
|
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
6M8Z
 
 | | Crystal structure of human DJ-1 without a modification on Cys-106 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Shumilin, I.A, Shabalin, I.G, Shumilina, S.V, Werenskjold, C, Utepbergenov, D, Minor, W. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A transient post-translational modification of active site cysteine alters binding properties of the parkinsonism protein DJ-1.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4OJJ
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
