5TQP
 
 | | LIPOXYGENASE-1 (SOYBEAN) I553G MUTANT AT 300K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-Deuterium Exchange of Lipoxygenase Uncovers a Relationship between Distal, Solvent Exposed Protein Motions and the Thermal Activation Barrier for Catalytic Proton-Coupled Electron Tunneling.
ACS Cent Sci, 3, 2017
|
|
5T5V
 
 | | LIPOXYGENASE-1 (SOYBEAN) AT 293K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen-Deuterium Exchange of Lipoxygenase Uncovers a Relationship between Distal, Solvent Exposed Protein Motions and the Thermal Activation Barrier for Catalytic Proton-Coupled Electron Tunneling.
ACS Cent Sci, 3, 2017
|
|
7QFI
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain I (aa 31-182) | | Descriptor: | CALCIUM ION, SlpX | | Authors: | Sagmeister, T, Damisch, E, Millan, C, Uson, I, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFK
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II, Co-Crystallization with HgCl2, Mutation Ser316Cys (aa 194-362) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFJ
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II (aa 194-362) | | Descriptor: | SlpX | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C, Baek, M, Read, R, Baker, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFG
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain III (aa 309-444) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, S-layer protein | | Authors: | Sagmeister, T, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLH
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 48-213) | | Descriptor: | PHOSPHATE ION, S-layer, SODIUM ION | | Authors: | Grininger, C, Sagmeister, T, Eder, E, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3II5
 
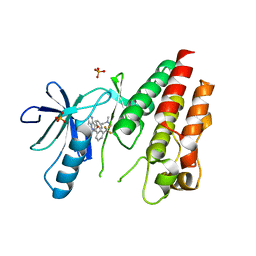 | | The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-[3-(3-{4-[(dimethylamino)methyl]phenyl}pyrazolo[1,5-a]pyrimidin-7-yl)phenyl]-3-(trifluoromethyl)benzamide, PHOSPHATE ION | | Authors: | Xu, W, Breger, D, Torres, N, Dutia, M, Powell, D, Ciszewski, G. | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Non-hinge-binding pyrazolo[1,5-a]pyrimidines as potent B-Raf kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3II4
 
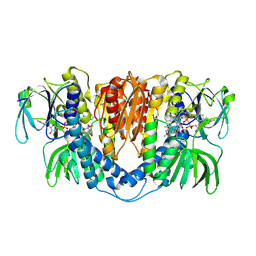 | |
5HI3
 
 | |
5HI4
 
 | | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists | | Descriptor: | (9'S,17'R)-6'-chloro-N-methyl-9'-{[(1-methyl-1H-pyrazol-5-yl)carbonyl]amino}-10',19'-dioxo-2'-oxa-11',18'-diazaspiro[cyclopentane-1,21'-tetracyclo[20.2.2.2~12,15~.1~3,7~]nonacosane]-1'(24'),3'(29'),4',6',12',14',22',25',27'-nonaene-17'-carboxamide, CAT-2000 FAB heavy chain, CAT-2000 FAB light chain, ... | | Authors: | Liu, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists.
Sci Rep, 6, 2016
|
|
1JN3
 
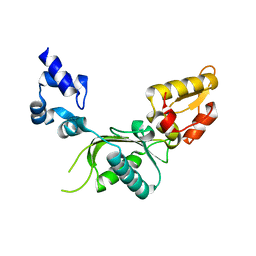 | |
5HI5
 
 | | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists | | Descriptor: | (4S,20R)-7-chloro-N-methyl-4-{[(1-methyl-1H-pyrazol-5-yl)carbonyl]amino}-3,18-dioxo-2,19-diazatetracyclo[20.2.2.1~6,10~.1~11,15~]octacosa-1(24),6(28),7,9,11(27),12,14,22,25-nonaene-20-carboxamide, CAT-2000 FAB heavy chain, CAT-2000 light chain, ... | | Authors: | Liu, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists.
Sci Rep, 6, 2016
|
|
2FDD
 
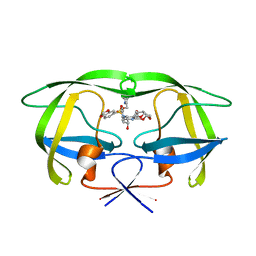 | | Crystal structure of HIV protease D545701 bound with GW0385 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-3-[(1,3-BENZODIOXOL-5-YLSULFONYL)(ISOBUTYL)AMINO]-2-HYDROXY-1-{4-[(2-METHYL-1,3-THIAZOL-4-YL)METHOXY]BENZYL}PROPYL]CARBAMATE, Gag-Pol polyprotein | | Authors: | Xu, R.X. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Ultra-potent P1 modified arylsulfonamide HIV protease inhibitors: The discovery of GW0385.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4JPH
 
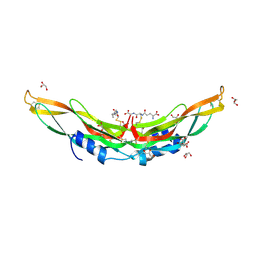 | | Crystal structure of Protein Related to DAN and Cerberus (PRDC) | | Descriptor: | CITRIC ACID, GLUTATHIONE, GLYCEROL, ... | | Authors: | Deng, X, Nolan, K.T, Kattamuri, C, Thompson, T.B. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of protein related to dan and cerberus: insights into the mechanism of bone morphogenetic protein antagonism.
Structure, 21, 2013
|
|
6TRI
 
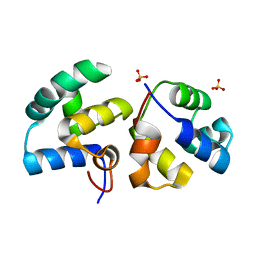 | | CI-MOR repressor-antirepressor complex of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | CI, MOR, SULFATE ION | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Jensen, M.R, Lo Leggio, L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TO6
 
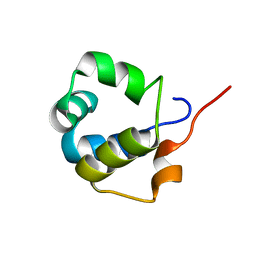 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | MOR | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7S13
 
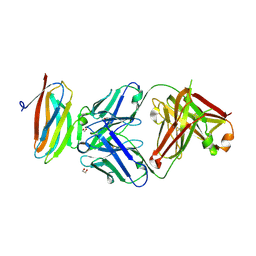 | | Crystal structure of Fab in complex with mouse CD96 dimer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Lee, P.S, Barman, I, Strop, P. | | Deposit date: | 2021-08-31 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Antibody blockade of CD96 by distinct molecular mechanisms.
Mabs, 13, 2021
|
|
7S11
 
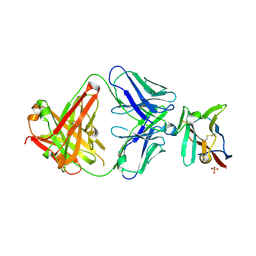 | | Crystal structure of Fab in complex with mouse CD96 monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-08-31 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Antibody blockade of CD96 by distinct molecular mechanisms.
Mabs, 13, 2021
|
|
7KHD
 
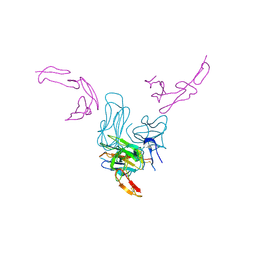 | | Human GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18 | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.956102 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
7KHX
 
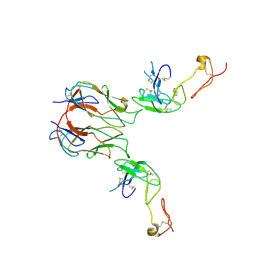 | | Mouse GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, ... | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.20569849 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
4CPG
 
 | |
7CBS
 
 | | Crystal structure of SpaB basal pilin from Lactobacillus rhamnosus GG | | Descriptor: | CHLORIDE ION, LPXTG cell wall anchor domain-containing protein, MAGNESIUM ION | | Authors: | Megta, A.K, Pratap, S, Kant, A, Krishnan, V. | | Deposit date: | 2020-06-13 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the atypically adhesive SpaB basal pilus subunit: Mechanistic insights about its incorporation in lactobacillar SpaCBA pili.
Curr Res Struct Biol, 2, 2020
|
|
7S7I
 
 | | Crystal structure of Fab in complex with MICA alpha3 domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Lee, P.S, Strop, P. | | Deposit date: | 2021-09-16 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residue-Level Characterization of Antibody Binding Epitopes Using Carbene Chemical Footprinting.
Anal.Chem., 95, 2023
|
|
7SU1
 
 | | Crystal structure of an acidic pH-selective Ipilimumab variant Ipi.106 in complex with CTLA-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, Fab heavy chain, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Improved therapeutic index of an acidic pH-selective antibody.
Mabs, 14, 2022
|
|
