7SU1
 
 | | Crystal structure of an acidic pH-selective Ipilimumab variant Ipi.106 in complex with CTLA-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, Fab heavy chain, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Improved therapeutic index of an acidic pH-selective antibody.
Mabs, 14, 2022
|
|
7SU0
 
 | | Crystal structure of an acidic pH-selective Ipilimumab variant Ipi.105 in complex with CTLA-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Cytotoxic T-lymphocyte protein 4, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Improved therapeutic index of an acidic pH-selective antibody.
Mabs, 14, 2022
|
|
7KHD
 
 | | Human GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18 | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.956102 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
7KHX
 
 | | Mouse GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, ... | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.20569849 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
4AIF
 
 | | AIP TPR domain in complex with human Hsp90 peptide | | Descriptor: | AH RECEPTOR-INTERACTING PROTEIN, HEAT SHOCK PROTEIN HSP 90-ALPHA, SULFATE ION | | Authors: | Morgan, R.M.L, Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2012-02-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structure of the Tpr Domain of Aip: Lack of Client Protein Interaction with the C-Terminal Alpha-7 Helix of the Tpr Domain of Aip is Sufficient for Pituitary Adenoma Predisposition.
Plos One, 7, 2012
|
|
4APO
 
 | | AIP TPR domain in complex with human Tomm20 peptide | | Descriptor: | AH RECEPTOR-INTERACTING PROTEIN, DODECAETHYLENE GLYCOL, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG, ... | | Authors: | Morgan, R.M.L, Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2012-04-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure of the Tpr Domain of Aip: Lack of Client Protein Interaction with the C-Terminal Alpha-7 Helix of the Tpr Domain of Aip is Sufficient for Pituitary Adenoma Predisposition.
Plos One, 7, 2012
|
|
4G10
 
 | | LigG from Sphingobium sp. SYK-6 is related to the glutathione transferase omega class | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase homolog, ... | | Authors: | Meux, E, Prosper, P, Masai, E, Mulliert Carlin, G, Dumarcay, S, Morel, M, Didierjean, C, Gelhaye, E, Favier, F. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sphingobium sp. SYK-6 LigG involved in lignin degradation is structurally and biochemically related to the glutathione transferase omega class.
Febs Lett., 586, 2012
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1H85
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH VAL 136 REPLACED BY LEU (V136L) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a Cluster of Hydrophobic Residues Near the Fad Cofactor in Anabaena Pcc 7119 Ferredoxin-Nadp+ Reductase for Optimal Complex Formation and Electron Transfer to Ferredoxin
J.Biol.Chem., 276, 2001
|
|
1H42
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH THR 155 REPLACED BY GLY, ALA 160 REPLACED BY THR AND LEU 263 REPLACED BY PRO (T155G-A160T-L263P) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2002-09-26 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Involvement of the Pyrophosphate and the 2'-Phosphate Binding Regions of Ferredoxin-Nadp+ Reductase in Coenzyme Specificity
J.Biol.Chem., 278, 2003
|
|
4L1Z
 
 | |
4L1Y
 
 | |
4L20
 
 | |
4L21
 
 | |
4OOF
 
 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203F mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
4OOE
 
 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203Y mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
1J7A
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN D68K MUTANT | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, VanHooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollen, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
3EFY
 
 | |
1OGJ
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 263 REPLACED BY PRO (L263P) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Martinez Julvez, M, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Involvement of the Pyrophosphate and the 2'-Phosphate Binding Regions of Ferredoxin-Nadp+ Reductase in Coenzyme Specificity
J.Biol.Chem., 278, 2003
|
|
1OGI
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH THR 155 REPLACED BY GLY AND ALA 160 REPLACED BY THR (T155G-A160T) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Julvez, M.M, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Involvement of the Pyrophosphate and the 2'-Phosphate Binding Regions of Ferredoxin-Nadp+ Reductase in Coenzyme Specificity.
J.Biol.Chem., 278, 2003
|
|
3GQM
 
 | |
1QOA
 
 | | FERREDOXIN MUTATION C49S | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Iron-sulfur cluster cysteine-to-serine mutants of Anabaena -2Fe-2S- ferredoxin exhibit unexpected redox properties and are competent in electron transfer to ferredoxin:NADP+ reductase.
Biochemistry, 36, 1997
|
|
1QGZ
 
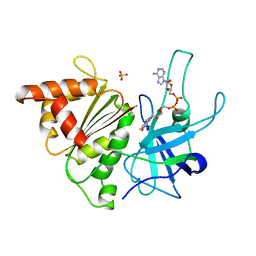 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 78 REPLACED BY ASP (L78D) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
3GQJ
 
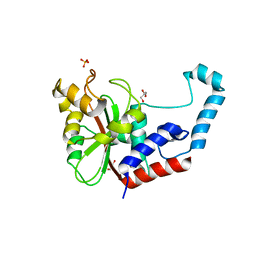 | |
1QOB
 
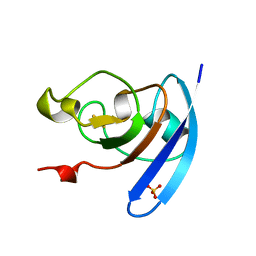 | | FERREDOXIN MUTATION D62K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
