1Z67
 
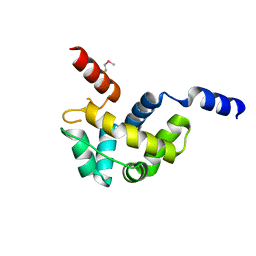 | | Structure of Homeodomain-like Protein of Unknown Function S4005 from Shigella flexneri | | Descriptor: | SODIUM ION, hypothetical protein S4005 | | Authors: | Osipiuk, J, Maltseva, N, Dementieva, I, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-21 | | Release date: | 2005-05-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of YidB protein from Shigella flexneri shows a new fold with homeodomain motif.
Proteins, 65, 2006
|
|
3MKL
 
 | |
3MR7
 
 | |
1KK0
 
 | |
2AHO
 
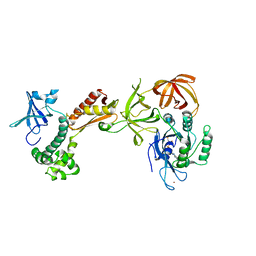 | | Structure of the archaeal initiation factor eIF2 alpha-gamma heterodimer from Sulfolobus solfataricus complexed with GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Translation initiation factor 2 alpha subunit, ... | | Authors: | Yatime, L, Mechulam, Y, Blanquet, S, Schmitt, E. | | Deposit date: | 2005-07-28 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Switch of the gamma Subunit in an Archaeal aIF2alphagamma Heterodimer
Structure, 14, 2006
|
|
3L0Z
 
 | |
3N2Q
 
 | |
3NE7
 
 | | Crystal structure of paia n-acetyltransferase from thermoplasma acidophilum in complex with coenzyme a | | Descriptor: | ACETYLTRANSFERASE, BETA-MERCAPTOETHANOL, COENZYME A, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
2HAY
 
 | | The Crystal Structure of the Putative NAD(P)H-Flavin Oxidoreductase from Streptococcus pyogenes M1 GAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative NAD(P)H-flavin oxidoreductase, SULFATE ION | | Authors: | Kim, Y, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-13 | | Release date: | 2006-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Crystal Structure of the Putative NAD(P)H-Flavin Oxidoreductase from Streptococcus pyogenes M1 GAS
To be Published, 2006
|
|
2HNG
 
 | | The Crystal Structure of Protein of Unknown Function SP1558 from Streptococcus pneumoniae | | Descriptor: | Hypothetical protein | | Authors: | Kim, Y, Zhang, D, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-12 | | Release date: | 2006-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Crystal Structure of Hypothetical Protein SP_1558 from Streptococcus pneumoniae
To be Published, 2006
|
|
2I1S
 
 | |
2I5E
 
 | | Crystal Structure of a Protein of Unknown Function MM2497 from Methanosarcina mazei Go1, Probable Nucleotidyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Hypothetical protein MM_2497 | | Authors: | Tan, K, Du, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a hypothetical protein MM_2497 from Methanosarcina mazei Go1
To be Published
|
|
6Q2B
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator from Listeria monocytogenes complexed with 26mer DNA | | Descriptor: | ACETIC ACID, DNA (26-MER), MarR family transcriptional regulator | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-08-07 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator from Listeria monocytogenes complexed with 26mer DNA.
To Be Published
|
|
3O5V
 
 | | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A | | Descriptor: | CHLORIDE ION, GLYCEROL, X-PRO dipeptidase | | Authors: | Stein, A.J, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Creatinase/Prolidase N-terminal domain of an X-PRO dipeptidase from Streptococcus pyogenes to 1.85A
To be Published
|
|
5L3E
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6,7-dimethoxyquinazoline-2,4-diamine, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-04-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5L3F
 
 | | LSD1-CoREST1 in complex with polymyxin B | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, Polmyxin B, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-04-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5L3G
 
 | | LSD1-CoREST1 in complex with polymyxin E (colistin) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-04-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5LBQ
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N2-(3-(dimethylamino)propyl)-6,7-dimethoxy-N4,N4-dimethylquinazoline-2,4-diamine, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
3OOS
 
 | | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne | | Descriptor: | Alpha/beta hydrolase family protein, GLYCEROL, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Bigelow, L, Hamilton, J, Li, H, Zhou, Y, Clancy, S, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne
To be Published
|
|
8B80
 
 | | The structure of Gan1D W433A in complex with galactose-6P | | Descriptor: | 6-O-phosphono-beta-D-galactopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Snyder, J, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2022-10-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of Gan1D W433A in complex with galactose-6P
To Be Published
|
|
8B81
 
 | | The structure of Gan1D W433A in complex with cellobiose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, IMIDAZOLE, Putative 6-phospho-beta-galactobiosidase | | Authors: | Snyder, J, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2022-10-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | The structure of Gan1D W433A in complex with cellobiose-6-phosphate
To Be Published
|
|
3PU9
 
 | | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein serine/threonine phosphatase | | Authors: | Nocek, B, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-22 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745
TO BE PUBLISHED
|
|
3PSL
 
 | | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3 based peptide mimetic | | Descriptor: | N-alpha acetylated form of histone H3, WD repeat-containing protein 5 | | Authors: | Avdic, V, Zhang, P, Lanouette, S, Voronova, A, Skerjanc, I, Couture, J.-F. | | Deposit date: | 2010-12-01 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3-based peptide mimetic.
Faseb J., 25, 2011
|
|
3OOO
 
 | | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V | | Descriptor: | Proline dipeptidase | | Authors: | Fan, Y, Wu, R, Morales, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V
To be Published
|
|
3OOV
 
 | | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287 | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, putative | | Authors: | Joachimiak, A, Duke, N.E.C, Hatzos-Skintges, C, Mulligan, R, Clancy, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287
To be Published
|
|
