9MHT
 
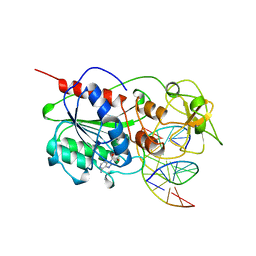 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*(3DR)P*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-07 | | Release date: | 1998-12-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
8UVL
 
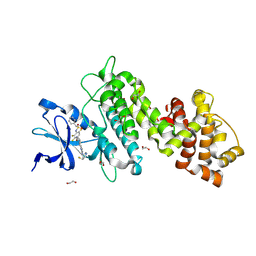 | | Crystal structure of selective IRE1a inhibitor 29 at the enzyme active site | | Descriptor: | 1,2-ETHANEDIOL, 1-phenyl-N-(2,3,6-trifluoro-4-{[(3M)-3-(2-{[(3R,5R)-5-fluoropiperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}phenyl)methanesulfonamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Kiefer, J.R, Wallweber, H.A, Braun, M.-G, Wei, W, Jiang, F, Wang, W, Rudolph, J, Ashkenazi, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Potent, Selective, and Orally Available IRE1 alpha Inhibitors Demonstrating Comparable PD Modulation to IRE1 Knockdown in a Multiple Myeloma Model.
J.Med.Chem., 67, 2024
|
|
8UTG
 
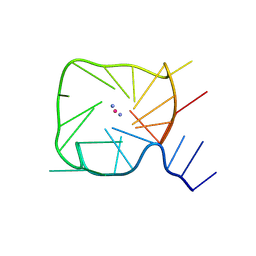 | | An RNA G-Quadruplex from the NS5 gene in the West Nile Virus Genome | | Descriptor: | AMMONIUM ION, POTASSIUM ION, RNA (5'-R(*UP*AP*UP*GP*GP*AP*AP*GP*AP*GP*GP*CP*GP*GP*UP*UP*GP*GP*UP*AP*U)-3') | | Authors: | Terrell, J.R, Le, T.T, Wilson, W.D, Siemer, J.L. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of an RNA G-quadruplex from the West Nile virus genome.
Nat Commun, 15, 2024
|
|
8XKL
 
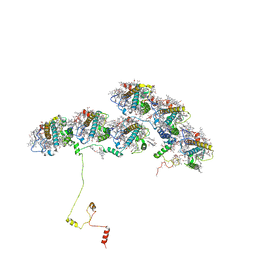 | | Structure of ACPII-CCPII from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
1RKM
 
 | | STRUCTURE OF OPPA | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
7ZVI
 
 | | Non-canonical Staphylococcus aureus pathogenicity island repression | | Descriptor: | Orf22, Sri | | Authors: | Miguel-Romero, L, Alqasmi, M, Bacarizo, J, Marina, A, Penades, J.R. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.973 Å) | | Cite: | Non-canonical Staphylococcus aureus pathogenicity island repression.
Nucleic Acids Res., 50, 2022
|
|
8A4F
 
 | | Human Interleukin-4 mutant - C3T-IL4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Mueller, T.D, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2022-06-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
8AQQ
 
 | |
8AQR
 
 | |
8BI8
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase | | Descriptor: | Nitric oxide synthase, brain | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
8BI9
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase (T112W/T116E variant) | | Descriptor: | Nitric oxide synthase, brain, TERTIARY-BUTYL ALCOHOL | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
8BVO
 
 | |
8BXR
 
 | |
8BW6
 
 | | Titin FnIII-domain I110 (I/A6) from the MIR region | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mayans, O, Fleming, J.R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Immunological and Structural Characterization of Titin Main Immunogenic Region; I110 Domain Is the Target of Titin Antibodies in Myasthenia Gravis.
Biomedicines, 11, 2023
|
|
8BCK
 
 | |
8BC5
 
 | |
8BBT
 
 | |
8BCL
 
 | |
8BS7
 
 | |
8A31
 
 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC694 | | Descriptor: | 4-(3-fluoranylpyrrol-1-yl)-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8A32
 
 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC769 | | Descriptor: | 1,2-ETHANEDIOL, 4-[3,4-bis(fluoranyl)pyrrol-1-yl]-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8BNQ
 
 | |
8C9M
 
 | | HERV-K Gag immature lattice | | Descriptor: | Gag protein | | Authors: | Krebs, A.-S, Liu, H.-F, Zhou, Y, Rey, J.S, Levintov, L, Perilla, J.R, Bartesaghi, A, Zhang, P. | | Deposit date: | 2023-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and conservation of an immature human endogenous retrovirus.
Biorxiv, 2023
|
|
5M0W
 
 | | N-terminal domain of mouse Shisa 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lohkamp, B, Ojala, J.R.M. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ab initio solution of macromolecular crystal structures without direct methods.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8IRG
 
 | | XFEL structure of cyanobacterial photosystem II following two flashes (2F) with a 30-microsecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
