8XUV
 
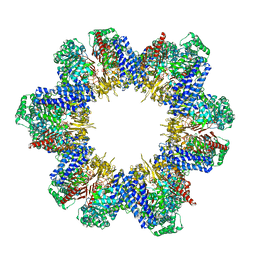 | | Cryo-EM structure of tomato NRC2 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
8XUO
 
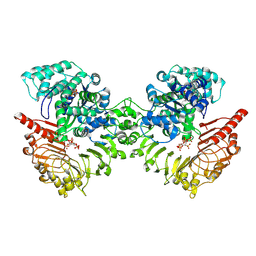 | | Cryo-EM structure of tomato NRC2 dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-13 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
8XUQ
 
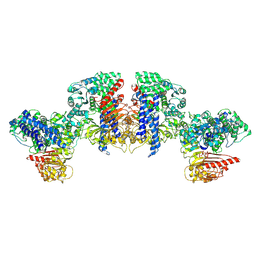 | | Cryo-EM structure of tomato NRC2 tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
6WHC
 
 | | CryoEM Structure of the glucagon receptor with a dual-agonist peptide | | Descriptor: | Dual-agonist peptide, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Sexton, P, Danev, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-electron microscopy structure of the glucagon receptor with a dual-agonist peptide.
J.Biol.Chem., 295, 2020
|
|
8JIP
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIR
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIT
 
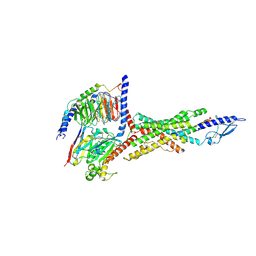 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIQ
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist Peptide 15-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIU
 
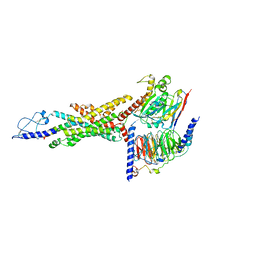 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIS
 
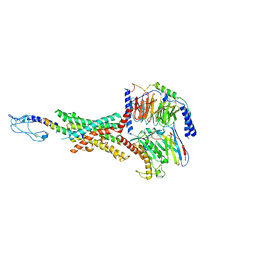 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist peptide15-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6L8V
 
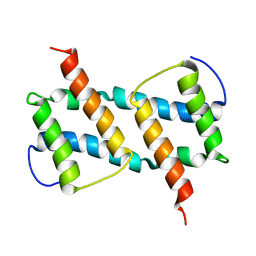 | | membrane-bound Bax helix2-helix5 domain | | Descriptor: | Apoptosis regulator BAX | | Authors: | OuYang, B, Lv, F. | | Deposit date: | 2019-11-07 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An amphipathic Bax core dimer forms part of the apoptotic pore wall in the mitochondrial membrane.
Embo J., 40, 2021
|
|
6MK4
 
 | |
2WF6
 
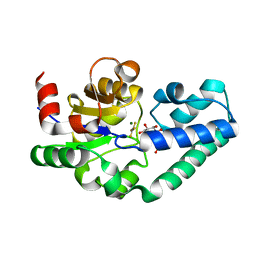 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6-phosphate and Aluminium tetrafluoride | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
2WF5
 
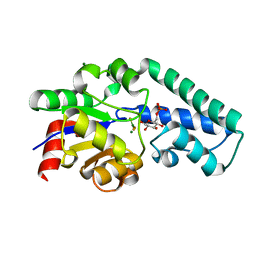 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6-phosphate and trifluoromagnesate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Hounslow, A.M, Cliff, M.J, Williams, N.H, Hollfelder, F, Gamblin, S, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
6MK5
 
 | |
2WHE
 
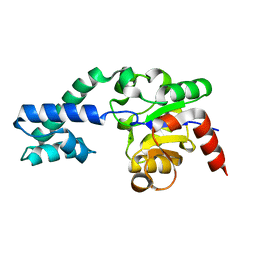 | | Structure of native Beta-Phosphoglucomutase in an open conformation without bound ligands. | | Descriptor: | BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic Details of Near-Transition State Conformers for Enzyme Phosphoryl Transfer Revealed by Mgf-3 Rather Than by Phosphoranes.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1J4I
 
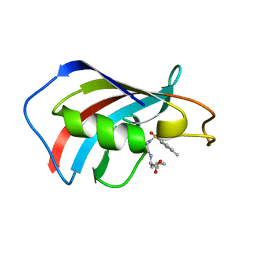 | | crystal structure analysis of the FKBP12 complexed with 000308 small molecule | | Descriptor: | 4-METHYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PENTANOIC ACID, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
1J4H
 
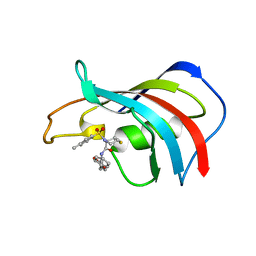 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | Descriptor: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
5YAD
 
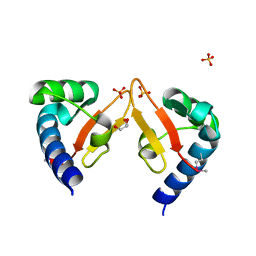 | | Crystal structure of Marf1 Lotus domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1, SULFATE ION | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6JNL
 
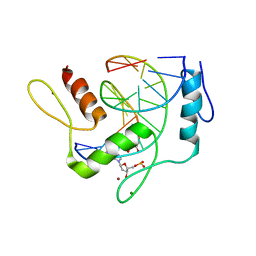 | | REF6 ZnF2-4-NAC004 complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
5YAA
 
 | | Crystal structure of Marf1 NYN domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1 | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7VSK
 
 | | Crystal structure of E.coli pseudouridine kinase PsuK complexed with pseudouridine. | | Descriptor: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, PfkB domain protein | | Authors: | Li, K.J, Li, X.J, Wu, B.X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure Characterization of Escherichia coli Pseudouridine Kinase PsuK.
Front Microbiol, 13, 2022
|
|
7VKP
 
 | |
6JNM
 
 | | REF6 ZnF2-4-NAC004-mC3 complex | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*(5CM)P*TP*GP*TP*TP*TP*TP*G)-3'), Lysine-specific demethylase REF6, ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
6JNN
 
 | | REF6 ZnF2-4-NAC004-mC1 complex | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*(5CM)P*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), Lysine-specific demethylase REF6, ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
