6JIV
 
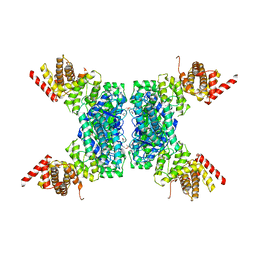 | | SspE crystal structure | | Descriptor: | SspE protein | | Authors: | Bing, Y.Z, Yang, H.G. | | Deposit date: | 2019-02-23 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
6JUF
 
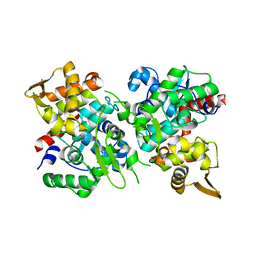 | | SspB crystal structure | | Descriptor: | SspB protein | | Authors: | Liqiong, L, Yubing, Z. | | Deposit date: | 2019-04-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
7V47
 
 | |
7V48
 
 | |
7V49
 
 | |
7V8E
 
 | | Crystal structure of IpaH1.4 LRR domain bound to HOIL-1L UBL domain. | | Descriptor: | RING-type E3 ubiquitin transferase, RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8G
 
 | | Crystal structure of HOIP RING1 domain bound to IpaH1.4 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, RING-type E3 ubiquitin transferase, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8F
 
 | | Crystal structure of UBE2L3 bound to HOIP RING1 domain. | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Ubiquitin-conjugating enzyme E2 L3, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8H
 
 | | Crystal structure of LRR domain from Shigella flexneri IpaH1.4 | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6L0X
 
 | | The First Tudor Domain of PHF20L1 | | Descriptor: | CITRIC ACID, GLYCEROL, PHD finger protein 20-like protein 1 | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1P
 
 | | Crystal structure of PHF20L1 in complex with Hit 1 | | Descriptor: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24L mutant | | Descriptor: | GLYCEROL, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1I
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24W/Y29W mutant | | Descriptor: | PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6KJG
 
 | | Crystal structure of PsoF | | Descriptor: | Dual-functional monooxygenase/methyltransferase psoF | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analyses oftrans C-Methyltransferase in Fungal Polyketide Biosynthesis.
Biochemistry, 58, 2019
|
|
6KJI
 
 | | Crystal structure of PsoF with SAH | | Descriptor: | Dual-functional monooxygenase/methyltransferase psoF, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analyses oftrans C-Methyltransferase in Fungal Polyketide Biosynthesis.
Biochemistry, 58, 2019
|
|
6KAW
 
 | | Crystal structure of CghA | | Descriptor: | CghA | | Authors: | Hara, K, Hashimoto, H, Yokoyama, M, Sato, M, Watanabe, K. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels-Alderase
Nat Catal, 2021
|
|
6KBC
 
 | | Crystal structure of CghA with Sch210972 | | Descriptor: | (2S)-3-[(2S,4E)-4-[[(1R,2S,4aR,6S,8R,8aS)-2-[(E)-but-2-en-2-yl]-6,8-dimethyl-1,2,4a,5,6,7,8,8a-octahydronaphthalen-1-yl]-oxidanyl-methylidene]-3,5-bis(oxidanylidene)pyrrolidin-2-yl]-2-methyl-2-oxidanyl-propanoic acid, CghA | | Authors: | Hara, K, Hashimoto, H, Maeda, N, Sato, M, Watanabe, K. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels-Alderase
Nat Catal, 2021
|
|
6L6W
 
 | | The structure of ScoE with intermediate | | Descriptor: | (3R)-3-[[(1R)-1,2-bis(oxidanyl)-2-oxidanylidene-ethyl]amino]butanoic acid, FE (II) ION, FORMIC ACID, ... | | Authors: | Chen, T.Y, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Pathway from N-Alkylglycine to Alkylisonitrile Catalyzed by Iron(II) and 2-Oxoglutarate-Dependent Oxygenases.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5XJ5
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the monoacylglycerol form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLYCINE, Glycerol-3-phosphate acyltransferase, ... | | Authors: | Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
3TL1
 
 | | Crystal structure of the Streptomyces coelicolor WhiE ORFVI polyketide aromatase/cyclase | | Descriptor: | 6,7,9-trihydroxy-3-methyl-1H-benzo[g]isochromen-1-one, GLYCEROL, Polyketide cyclase | | Authors: | Lee, M.-Y, Ames, B.D, Zhang, W, Tang, Y, Tsai, S.-C. | | Deposit date: | 2011-08-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the Molecular Basis of Aromatic Polyketide Cyclization: Crystal Structure and in Vitro Characterization of WhiE-ORFVI.
Biochemistry, 51, 2012
|
|
5YY2
 
 | | Crystal structure of AsqI with Zn | | Descriptor: | Uncharacterized protein AsqI, ZINC ION | | Authors: | Hara, K, Hashimoto, H, Kishimoto, S, Watanabe, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Enzymatic one-step ring contraction for quinolone biosynthesis.
Nat Commun, 9, 2018
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
5YY3
 
 | |
8JI8
 
 | | Crystal Structure of Prophenoloxidase PPO6 chimeric mutant (F215EASNRAIVD224 to G215DGPDSVVR223) from Aedes aegypti | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, ... | | Authors: | Zhu, X, Zhang, L, Yang, X, Bao, P, Ren, D, Han, Q. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mosquitoes have evolved two types of prophenoloxidases
To Be Published
|
|
7WKC
 
 | |
