6O7I
 
 | |
6O6V
 
 | |
6O73
 
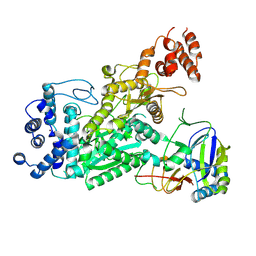 | | Crystal structure of apo Csm1-Csm4 cassette | | Descriptor: | Csm1, Csm4, NICKEL (II) ION | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
1A8W
 
 | |
177D
 
 | |
6O6X
 
 | |
1AFF
 
 | | DNA QUADRUPLEX CONTAINING GGGG TETRADS AND (T.A).A TRIADS, NMR, 8 STRUCTURES | | Descriptor: | QUADRUPLEX DNA (5'-D(TP*AP*GP*G)-3') | | Authors: | Kettani, A, Bouaziz, S, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-03-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads.
Nat.Struct.Biol., 4, 1997
|
|
193D
 
 | |
1A4T
 
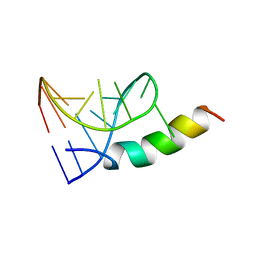 | | SOLUTION STRUCTURE OF PHAGE P22 N PEPTIDE-BOX B RNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | 20-MER BASIC PEPTIDE, BOXB RNA | | Authors: | Cai, Z, Gorin, A.A, Frederick, R, Ye, X, Hu, W, Majumdar, A, Kettani, A, Patel, D.J. | | Deposit date: | 1998-02-04 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex.
Nat.Struct.Biol., 5, 1998
|
|
1A6H
 
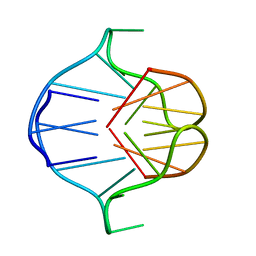 | |
179D
 
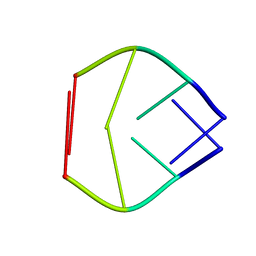 | |
186D
 
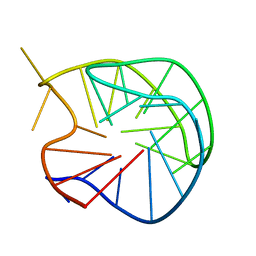 | |
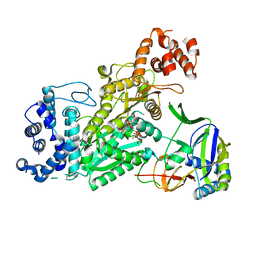
6O74
 
 | |
1AM0
 
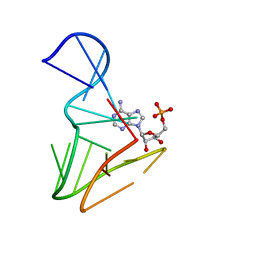 | | AMP RNA APTAMER COMPLEX, NMR, 8 STRUCTURES | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA APTAMER | | Authors: | Jiang, F, Kumar, R.A, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-06-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of RNA Folding and Recognition in an AMP-RNA Aptamer Complex
Nature, 382, 1996
|
|
6O7D
 
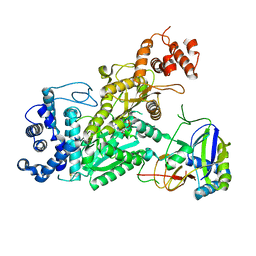 | | Crystal structure of Csm1-Csm4 cassette in complex with one ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
199D
 
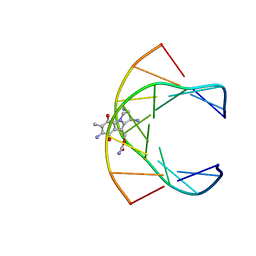 | | Solution structure of the monoalkylated mitomycin c-DNA complex | | Descriptor: | CARBAMIC ACID 2,6-DIAMINO-5-METHYL-4,7-DIOXO-2,3,4,7-TETRAHYDRO-1H-3A-AZA-CYCLOPENTA[A]INDEN-8-YLMETHYL ESTER, DNA (5'-D(*(DI)P*CP*AP*CP*GP*TP*CP*(DI)P*T)-3'), DNA (5'-D(*AP*CP*GP*AP*CP*GP*TP*GP*C)-3') | | Authors: | Sastry, M, Fiala, R, Lipman, R, Tomasz, M, Patel, D.J. | | Deposit date: | 1994-12-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monoalkylated mitomycin C-DNA complex.
J.Mol.Biol., 247, 1995
|
|
1A8N
 
 | | SOLUTION STRUCTURE OF A NA+ CATION STABILIZED DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS FORMED BY G-G-G-C REPEATS OBSERVED IN AAV AND HUMAN CHROMOSOME 19, NMR, 8 STRUCTURES | | Descriptor: | DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS | | Authors: | Kettani, A, Bouaziz, S, Gorin, A, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA.
J.Mol.Biol., 282, 1998
|
|
6O6S
 
 | | Crystal structure of Apo Csm6 | | Descriptor: | Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O7H
 
 | |
6O70
 
 | | Crystal structure of Csm6 H132A mutant in complex with cA4 by cocrystallization of cA4 and Csm6 H132A mutant | | Descriptor: | 2',3'- cyclic AMP, 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O6Z
 
 | | Crystal structure of Csm6 H381A in complex with cA4 by cocrystallization of cA4 and Csm6 | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O79
 
 | | Crystal structure of Csm1-Csm4 cassette in complex with cA3 | | Descriptor: | CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, cyclic RNA cA3 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
1B3P
 
 | | 5'-D(*GP*GP*AP*GP*GP*AP*T)-3' | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*AP*T)-3') | | Authors: | Kettani, A, Bouaziz, S, Skripkin, E, Majumdar, A, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-12-14 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Interlocked mismatch-aligned arrowhead DNA motifs.
Structure Fold.Des., 7, 1999
|
|
6D8A
 
 | | RsAgo Ternary Complex with guide RNA and Target DNA Containing A-A Bulge Within the Seed Segment of the Target Strand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*AP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
1AXU
 
 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | Descriptor: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | Authors: | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
