8HPK
 
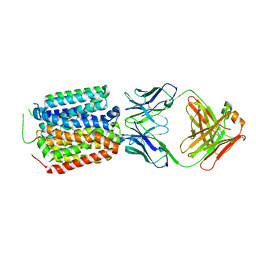 | | Crystal structure of the bacterial oxalate transporter OxlT in an oxalate-bound occluded form | | Descriptor: | Fab fragment Heavy chein, Fab fragment Light chain, OXALATE ION, ... | | Authors: | Shimamura, T, Hirai, T, Yamashita, A. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota.
Nat Commun, 14, 2023
|
|
8HPJ
 
 | |
4KRU
 
 | |
4KRT
 
 | |
6FDG
 
 | |
7JI2
 
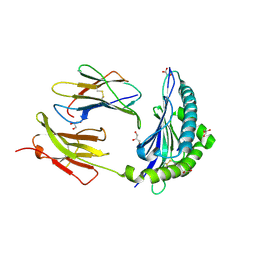 | | Crystal Structure of H2-Kb in complex with a OVA mutant peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
7JUK
 
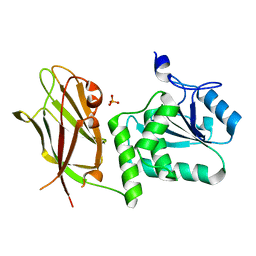 | | Crystal structure of PTEN with a tetra-phosphorylated tail (4p-crPTEN-13sp-T2, SDTTDSDPENEG) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JVX
 
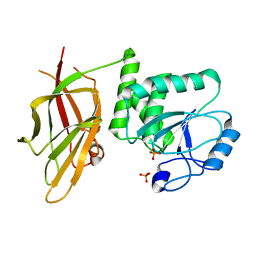 | | Crystal structure of PTEN (aa 7-353 followed by spacer TGGGSGGTGGGSGGTGGGCY ligated to peptide pSDpTpTDpSDPENEPFDED) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JUL
 
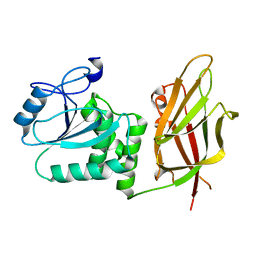 | | Crystal structure of non phosphorylated PTEN (n-crPTEN-13sp-T1, SDTTDSDPENEG) | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-20 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTX
 
 | |
6MF8
 
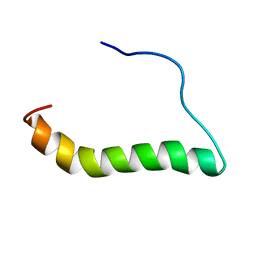 | | TCR alpha transmembrane domain | | Descriptor: | T-cell receptor alpha chain C region | | Authors: | Brazin, K.N, Reinherz, E.L. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The T Cell Antigen Receptor alpha Transmembrane Domain Coordinates Triggering through Regulation of Bilayer Immersion and CD3 Subunit Associations.
Immunity, 49, 2018
|
|
1NAS
 
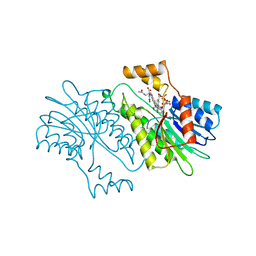 | | SEPIAPTERIN REDUCTASE COMPLEXED WITH N-ACETYL SEROTONIN | | Descriptor: | N-ACETYL SEROTONIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALOACETATE ION, ... | | Authors: | Auerbach, G, Herrmann, A, Bacher, A, Huber, R. | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
1IAC
 
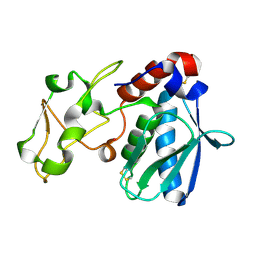 | | REFINED 1.8 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF ASTACIN, A ZINC-ENDOPEPTIDASE FROM THE CRAYFISH ASTACUS ASTACUS L. STRUCTURE DETERMINATION, REFINEMENT, MOLECULAR STRUCTURE AND COMPARISON WITH THERMOLYSIN | | Descriptor: | ASTACIN, MERCURY (II) ION | | Authors: | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined 1.8 A X-ray crystal structure of astacin, a zinc-endopeptidase from the crayfish Astacus astacus L. Structure determination, refinement, molecular structure and comparison with thermolysin.
J.Mol.Biol., 229, 1993
|
|
2RVH
 
 | | NMR structure of eIF1 | | Descriptor: | Eukaryotic translation initiation factor eIF-1 | | Authors: | Nagata, T, Obayashi, E, Asano, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5
Cell Rep, 18, 2017
|
|
4TQC
 
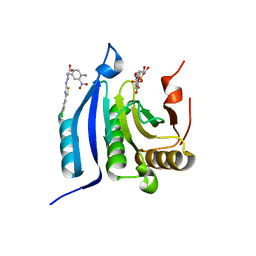 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2S)-3-(4-amino-3-nitrophenyl)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinyl}propanoic acid, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TPW
 
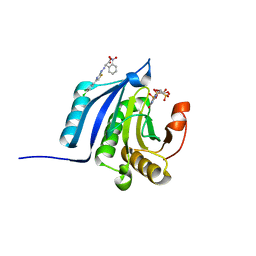 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TQB
 
 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(4-bromophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3A08
 
 | | Structure of (PPG)4-OOG-(PPG)4, monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Wu, G, Noguchi, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-03-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Two crystal modifications of (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 reveal the puckering preference of Hyp(X) in the Hyp(X):Hyp(Y) and Hyp(X):Pro(Y) stacking pairs in collagen helices.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A19
 
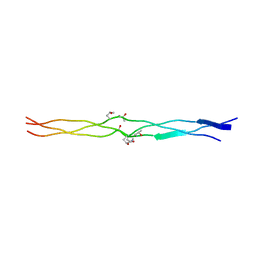 | | Structure of (PPG)4-OOG-(PPG)4_H monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Hyakutake, M, Wu, G, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-03-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two crystal modifications of (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 reveal the puckering preference of Hyp(X) in the Hyp(X):Hyp(Y) and Hyp(X):Pro(Y) stacking pairs in collagen helices.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2DRX
 
 | | Structure Analysis of (POG)4-(LOG)2-(POG)4 | | Descriptor: | collagen like peptide | | Authors: | Okuyama, K. | | Deposit date: | 2006-06-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unique side chain conformation of a leu residue in a triple-helical structure
Biopolymers, 86, 2007
|
|
2DRT
 
 | | Structure Analysis of (POG)4-LOG-(POG)5 | | Descriptor: | ETHANOL, collagen like peptide | | Authors: | Okuyama, K. | | Deposit date: | 2006-06-14 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unique side chain conformation of a leu residue in a triple-helical structure
Biopolymers, 86, 2007
|
|
1V4F
 
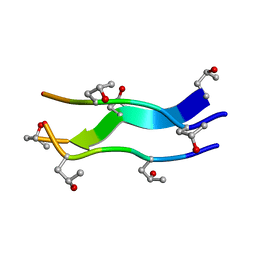 | | Crystal structures of collagen model peptides with pro-hyp-gly sequence at 1.3A | | Descriptor: | collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-11-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
2E9M
 
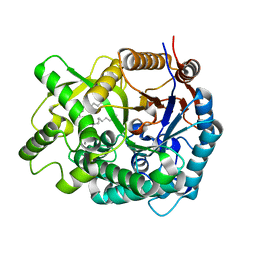 | | Crystal Structure of human Cytosolic Neutral beta-Glycosylceramidase (Klotho-related Prote:KLrP) complex with Galactose and fatty acids | | Descriptor: | Cytosolic beta-glucosidase, OLEIC ACID, PALMITIC ACID, ... | | Authors: | Kakuta, Y, Hayashi, Y, Okino, N, Ito, M. | | Deposit date: | 2007-01-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Klotho-related protein is a novel cytosolic neutral beta-glycosylceramidase.
J.Biol.Chem., 282, 2007
|
|
2E9L
 
 | | Crystal Structure of human Cytosolic Neutral beta-Glycosylceramidase (Klotho-related Prote:KLrP) complex with Glucose and fatty acids | | Descriptor: | Cytosolic beta-glucosidase, GLYCEROL, OLEIC ACID, ... | | Authors: | Kakuta, Y, Hayashi, Y, Okino, N, Ito, M. | | Deposit date: | 2007-01-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Klotho-related protein is a novel cytosolic neutral beta-glycosylceramidase.
J.Biol.Chem., 282, 2007
|
|
1GN7
 
 | |
