4CN1
 
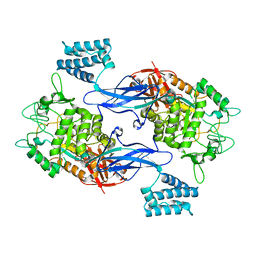 | | GlgE isoform 1 from Streptomyces coelicolor D394A mutant with maltose- 1-phosphate bound | | Descriptor: | ALPHA-1,4-GLUCAN: MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-1-O-phosphono-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
4CQ6
 
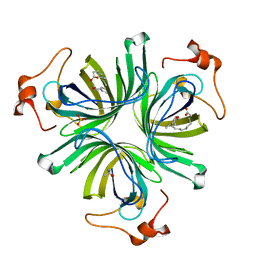 | | The crystal structure of the allene oxide cyclase 2 from Arabidopsis thaliana with bound inhibitor - vernolic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (9Z)-11-[(2R,3S)-3-pentyloxiran-2-yl]undec-9-enoic acid, ALLENE OXIDE CYCLASE 2, ... | | Authors: | Terlecka, B.A, Pollmann, S, Hofmann, E. | | Deposit date: | 2014-02-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Ligand-Bound Structures of the Aoc2 from A. Thaliana and the Implications for the Catalytic Mechanism
To be Published
|
|
4CN4
 
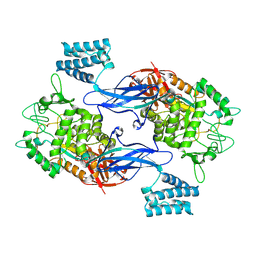 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant with 2-deoxy- 2-fluoro-beta-maltosyl modification | | Descriptor: | ALPHA-1,4-GLUCAN:MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-beta-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
4CQ7
 
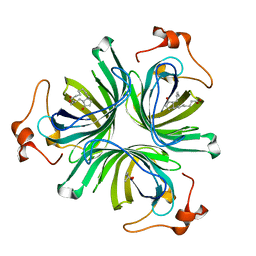 | | The crystal structure of the allene oxide cyclase 2 from Arabidopsis thaliana with bound product - OPDA | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 9S,13R-12-OXOPHYTODIENOIC ACID, ALLENE OXIDE CYCLASE 2, ... | | Authors: | Terlecka, B.A, Pollmann, S, Hofmann, E. | | Deposit date: | 2014-02-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Ligand-Bound Structures of the Aoc2 from A. Thaliana and the Implications for the Catalytic Mechanism
To be Published
|
|
7RGF
 
 | | Protocadherin gammaC4 EC1-4 crystal structure disrupted trans interface | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition.
Elife, 11, 2022
|
|
8DJH
 
 | | Ternary complex of SUMO1 with a phosphomimetic SIM of PML and zinc | | Descriptor: | PML 4SD, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
8DJI
 
 | | Ternary complex of SUMO1 with the SIM of PML and zinc | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
4BQE
 
 | | Arabidopsis thaliana Cytosolic Alpha-1,4-glucan Phosphorylase (PHS2) | | Descriptor: | ALPHA-GLUCAN PHOSPHORYLASE 2,4-GLUCAN PHOSPHORYLASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | O'Neill, E.C, Rashid, A.M, Stevenson, C.E.M, Hetru, A.C, Gunning, A.P, Rejzek, M, Nepogodiev, S.A, Bornemann, S, Lawson, D.M, Field, R.A. | | Deposit date: | 2013-05-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sugar-Coated Sensor Chip and Nanoparticle Surfaces for the in Vitro Enzymatic Synthesis of Starch-Like Materials
Chem.Sci., 5, 2014
|
|
6TX0
 
 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dAMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6E6B
 
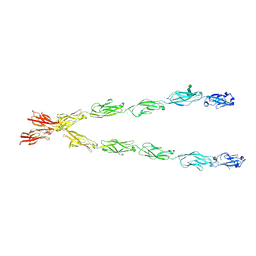 | | Crystal structure of the Protocadherin GammaB4 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2018-07-24 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.52 Å) | | Cite: | Visualization of clustered protocadherin neuronal self-recognition complexes.
Nature, 569, 2019
|
|
6TXE
 
 | | Crystal structure of tetrameric human wt-SAMHD1 (residues 109-626) with GTP, dATP, dTMPNPP and Mg | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6TXA
 
 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dGMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6TXC
 
 | | Crystal structure of tetrameric human wt-SAMHD1 (residues 109-626) with GTP, dATP, dCMPNPP and Mg | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6TVP
 
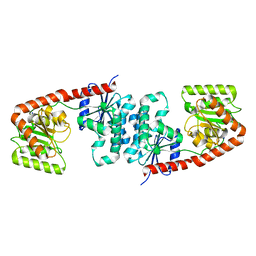 | | Structure of Mycobacterium smegmatis alpha-maltose-1-phosphate synthase GlgM | | Descriptor: | Alpha-maltose-1-phosphate synthase, SODIUM ION | | Authors: | Syson, K, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2020-01-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Mycobacterium smegmatis alpha-maltose-1-phosphate synthase GlgM.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TXF
 
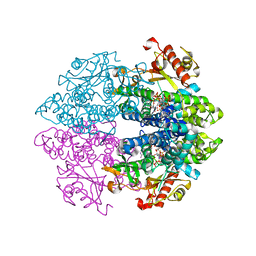 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dAMPNPP and Mn | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
4BQI
 
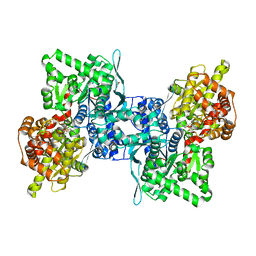 | | ARABIDOPSIS THALIANA cytosolic alpha-1,4-glucan phosphorylase (PHS2) in complex with maltotriose | | Descriptor: | ALPHA-GLUCAN PHOSPHORYLASE 2, CYTOSOLIC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | O'Neill, E.C, Rashid, A.M, Stevenson, C.E.M, Hetru, A.C, Gunning, A.P, Rejzek, M, Nepogodiev, S.A, Bornemann, S, Lawson, D.M, Field, R.A. | | Deposit date: | 2013-05-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sugar-Coated Sensor Chip and Nanoparticle Surfaces for the in Vitro Enzymatic Synthesis of Starch-Like Materials
Chem.Sci., 5, 2014
|
|
4BQF
 
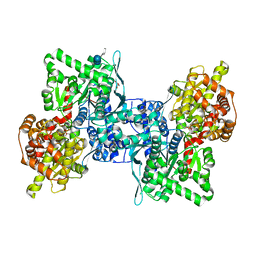 | | Arabidopsis thaliana cytosolic alpha-1,4-glucan phosphorylase (PHS2) in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-GLUCAN PHOSPHORYLASE 2, CYTOSOLIC, ... | | Authors: | O'Neill, E.C, Rashid, A.M, Stevenson, C.E.M, Hetru, A.C, Gunning, A.P, Rejzek, M, Nepogodiev, S.A, Bornemann, S, Lawson, D.M, Field, R.A. | | Deposit date: | 2013-05-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Sugar-Coated Sensor Chip and Nanoparticle Surfaces for the in Vitro Enzymatic Synthesis of Starch-Like Materials
Chem.Sci., 5, 2014
|
|
5DZX
 
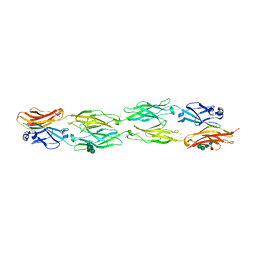 | | Protocadherin beta 6 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin beta 6, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5DZY
 
 | | Protocadherin beta 8 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Pcdhb8 protein, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5DZW
 
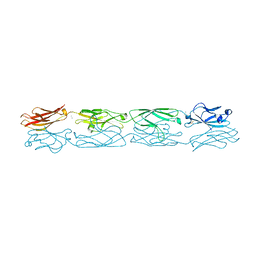 | | Protocadherin alpha 4 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin alpha-4, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5SZQ
 
 | | Protocadherin Gamma A4 extracellular cadherin domains 3-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5SZN
 
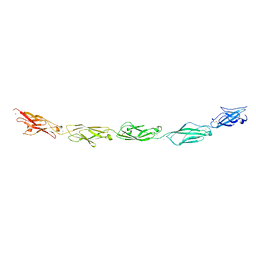 | | Protocadherin gamma A9 extracellular cadherin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.944 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5SZP
 
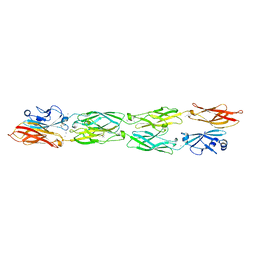 | | Protocadherin Gamma B7 extracellular cadherin domains 1-4 P21 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin Gamma B7, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5SZO
 
 | | Protocadherin Gamma B7 extracellular cadherin domains 1-4 P41212 crystal form | | Descriptor: | CALCIUM ION, Protocadherin Gamma B7, alpha-D-mannopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.612 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
7X7G
 
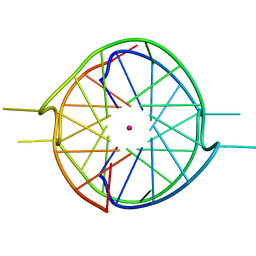 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
