8EPL
 
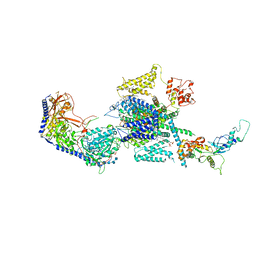 | | Human R-type voltage-gated calcium channel Cav2.3 at 3.1 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-10-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the R-type human Ca v 2.3 channel reveal conformational crosstalk of the intracellular segments.
Nat Commun, 13, 2022
|
|
8EPM
 
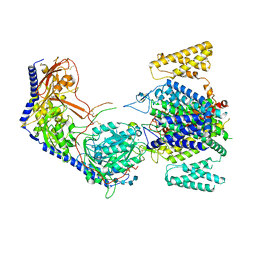 | | Human R-type voltage-gated calcium channel Cav2.3 CH2II-deleted mutant at 3.1 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-10-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the R-type human Ca v 2.3 channel reveal conformational crosstalk of the intracellular segments.
Nat Commun, 13, 2022
|
|
8FEZ
 
 | |
5IL1
 
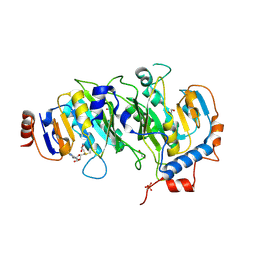 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL0
 
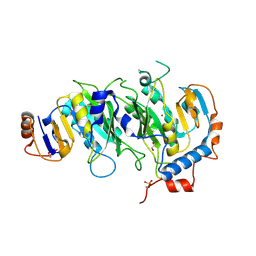 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL2
 
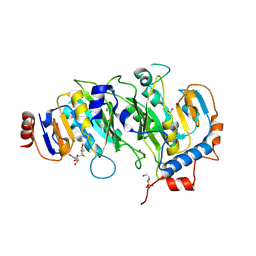 | | Crystal structure of SAH-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
8FBX
 
 | | Selenosugar synthase SenB from Variovorax paradoxus | | Descriptor: | CHLORIDE ION, SODIUM ION, Selenosugar synthase SenB | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2022-11-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Characterization and Ligand-Induced Conformational Changes of SenB, a Se-Glycosyltransferase Involved in Selenoneine Biosynthesis.
Biochemistry, 62, 2023
|
|
8FRZ
 
 | |
8FSB
 
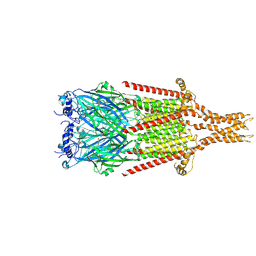 | |
8FSP
 
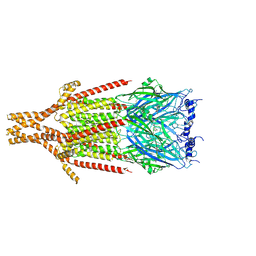 | | Full-length mouse 5-HT3A receptor in complex with SMP100, open-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FSZ
 
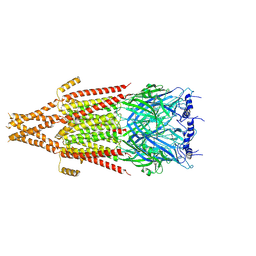 | | Full-length mouse 5-HT3A receptor in complex with ALB148471, open-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,5R)-1-azabicyclo[3.2.2]nonan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FRW
 
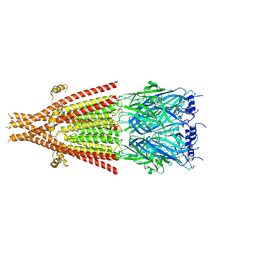 | | Full-length mouse 5-HT3A receptor in complex with ALB148471, pre-activated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,5R)-1-azabicyclo[3.2.2]nonan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-09 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FRX
 
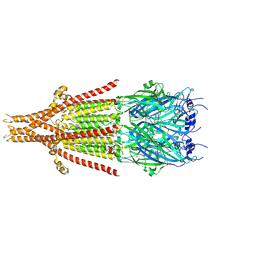 | | Full-length mouse 5-HT3A receptor in complex with SMP100, pre-activated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-09 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5JO5
 
 | |
5JNY
 
 | | Crystal Structure of 10E8 Fab | | Descriptor: | 10E8 Heavy Chain, 10E8 Light Chain, CHLORIDE ION, ... | | Authors: | Ofek, G, Kwong, P. | | Deposit date: | 2016-05-01 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8.
Plos One, 11, 2016
|
|
7SP0
 
 | | Crystal structure of human SFPQ L534I mutant in complex with zinc | | Descriptor: | Splicing factor, proline- and glutamine-rich, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2021-11-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Familial ALS-associated SFPQ variants promote the formation of SFPQ cytoplasmic aggregates in primary neurons.
Open Biology, 12, 2022
|
|
5JR1
 
 | |
6LI9
 
 | | Heteromeric amino acid transporter b0,+AT-rBAT complex bound with Arginine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
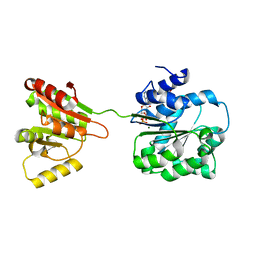 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
8UWA
 
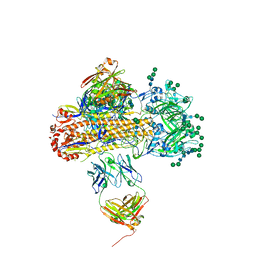 | | VH1-18 QxxV class antibody 09-1B12 bound to A/Perth/16/2009 H3N2 hemagglutinin | | Descriptor: | 09-1B12 heavy chain, 09-1B12 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maurer, D.P. | | Deposit date: | 2023-11-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.02 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
7MEW
 
 | | E. coli MsbA in complex with G247 | | Descriptor: | (2E)-3-{1-cyclopropyl-7-[(1S)-1-(3,6-dichloro-2-fluorophenyl)ethoxy]naphthalen-2-yl}prop-2-enoic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
7MET
 
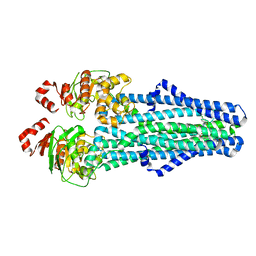 | | A. baumannii MsbA in complex with TBT1 decoupler | | Descriptor: | 2-(4-chlorobenzamido)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
4MCC
 
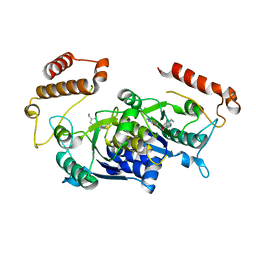 | | HinTrmD in complex with N-[4-(AMINOMETHYL)BENZYL]-4-OXO-3,4-DIHYDROTHIENO[2,3-D]PYRIMIDINE-5-CARBOXAMIDE | | Descriptor: | N-[4-(aminomethyl)benzyl]-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Olivier, N.B, Hill, P. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
