3G4M
 
 | |
4XYK
 
 | | Crystal structure of human phosphofructokinase-1 in complex with ADP, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
3GES
 
 | |
3GER
 
 | | Guanine riboswitch bound to 6-chloroguanine | | Descriptor: | 6-chloroguanine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Batey, R.T. | | Deposit date: | 2009-02-25 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs
Structure, 17, 2009
|
|
3GAO
 
 | |
4YMC
 
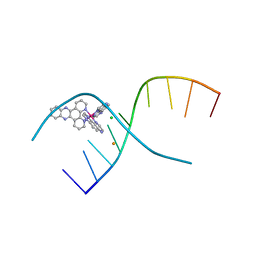 | | Lambda-[Ru(TAP)2(dppz)]2+ bound to d(CCGGATCCGG)2 | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*GP*AP*TP*CP*CP*GP*G)-3'), ... | | Authors: | Gurung, S.P, Hall, J.P, Cardin, C.J. | | Deposit date: | 2015-03-06 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Reversal of a single base pair step controls guanine photo-oxidation by an intercalating Ru(II) dipyridophenazine complex.
To Be Published
|
|
3GOG
 
 | |
5K2F
 
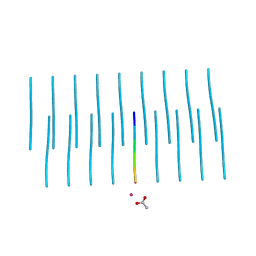 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1I2Y
 
 | | 1.66 A STRUCTURE OF A-DUPLEX WITH BULGED ADENOSINE, SPERMINE FORM | | Descriptor: | DNA/RNA (5'-R(*GP*CP*G)-D(P*AP*TP*AP*T)-R(P*AP*CP*GP*U)-3'), SPERMINE | | Authors: | Tereshko, V, Wallace, S, Usman, N, Wincott, F, Egli, M. | | Deposit date: | 2001-02-12 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | X-ray crystallographic observation of "in-line" and "adjacent" conformations in a bulged self-cleaving RNA/DNA hybrid.
RNA, 7, 2001
|
|
1I2X
 
 | | 2.4 A STRUCTURE OF A-DUPLEX WITH BULGED ADENOSINE, SPERMIDINE FORM | | Descriptor: | DNA/RNA (5'-R(*GP*CP*G)-D(P*AP*TP*AP*T)-R(P*AP*CP*GP*U)-3'), SPERMIDINE | | Authors: | Tereshko, V, Wallace, S, Usman, N, Wincott, F, Egli, M. | | Deposit date: | 2001-02-12 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic observation of "in-line" and "adjacent" conformations in a bulged self-cleaving RNA/DNA hybrid.
RNA, 7, 2001
|
|
5K2G
 
 | |
5KOJ
 
 | | Nitrogenase MoFeP protein in the IDS oxidized state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Owens, C.P, Tezcan, F.A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Tyrosine-Coordinated P-Cluster in G. diazotrophicus Nitrogenase: Evidence for the Importance of O-Based Ligands in Conformationally Gated Electron Transfer.
J.Am.Chem.Soc., 138, 2016
|
|
5KZS
 
 | | Listeria monocytogenes internalin-like protein lmo2027 | | Descriptor: | Putative cell surface protein, similar to internalin proteins | | Authors: | Light, S.H, Nocadello, S, Minasov, G, Cardona-Correa, A, Kwon, K, Faralla, C, Bakardjiev, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing.
Plos Pathog., 14, 2018
|
|
2FYY
 
 | |
2FZ3
 
 | |
2F9Q
 
 | | Crystal Structure of Human Cytochrome P450 2D6 | | Descriptor: | Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Rowland, P. | | Deposit date: | 2005-12-06 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Crystal Structure of Human Cytochrome P450 2D6
J.Biol.Chem., 281, 2006
|
|
2F40
 
 | |
5KOH
 
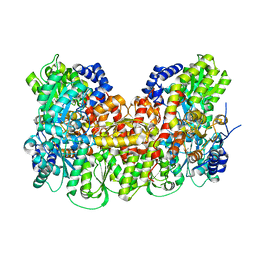 | | Nitrogenase MoFeP from Gluconacetobacter diazotrophicus in dithionite reduced state | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Owens, C.P, Tezcan, F.A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Tyrosine-Coordinated P-Cluster in G. diazotrophicus Nitrogenase: Evidence for the Importance of O-Based Ligands in Conformationally Gated Electron Transfer.
J.Am.Chem.Soc., 138, 2016
|
|
2GMG
 
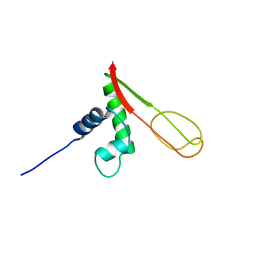 | | Solution NMR Structure of protein PF0610 from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfG3 | | Descriptor: | hypothetical protein Pf0610 | | Authors: | Wang, X, Lee, H.S, Adams, M.W, Northeast Structural Genomics Consortium (NESG), Montelione, G.T, Prestegard, J.H. | | Deposit date: | 2006-04-06 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PF0610, a novel winged helix-turn-helix variant possessing a rubredoxin-like Zn ribbon motif from the hyperthermophilic archaeon, Pyrococcus furiosus.
Biochemistry, 46, 2007
|
|
2HI3
 
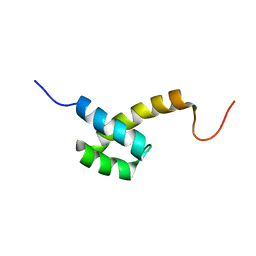 | | Solution structure of the homeodomain-only protein HOP | | Descriptor: | Homeodomain-only protein | | Authors: | Mackay, J.P, Kook, H, Epstein, J.A, Simpson, R.J, Yung, W.W. | | Deposit date: | 2006-06-28 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the structure and function of the transcriptional coregulator HOP
Biochemistry, 45, 2006
|
|
2HH0
 
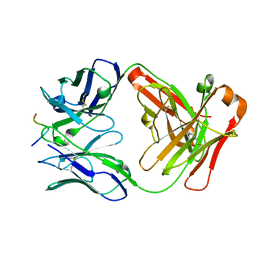 | |
4MRW
 
 | | Crystal structure of PDE10A2 with fragment ZT0120 (7-chloroquinolin-4-ol) | | Descriptor: | 7-chloroquinolin-4-ol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, NIenaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSH
 
 | | Crystal Structure of PDE10A2 with fragment ZT0143 ((2S)-4-chloro-2,3-dihydro-1,3-benzothiazol-2-amine) | | Descriptor: | 4-chloro-1,3-benzothiazol-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSC
 
 | | Crystal structure of PDE10A2 with fragment ZT1595 (2-[(quinolin-7-yloxy)methyl]quinoline) | | Descriptor: | 2-[(quinolin-7-yloxy)methyl]quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
