5D6T
 
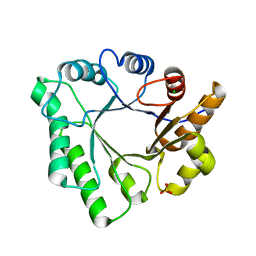 | | Crystal Structure of Aspergillus clavatus Sph3 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, SPHERULIN-4, ... | | Authors: | Bamford, N.C, Little, D.J, Howell, P.L. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Sph3 Is a Glycoside Hydrolase Required for the Biosynthesis of Galactosaminogalactan in Aspergillus fumigatus.
J.Biol.Chem., 290, 2015
|
|
5DA7
 
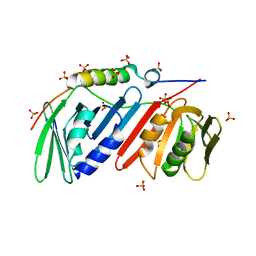 | | monomeric PCNA bound to a small protein inhibitor | | Descriptor: | DNA polymerase sliding clamp 1, Proliferating cell nuclear antigen, SULFATE ION, ... | | Authors: | Ladner, J.E, Altieri, A.S, Kelman, Z. | | Deposit date: | 2015-08-19 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A small protein inhibits proliferating cell nuclear antigen by breaking the DNA clamp.
Nucleic Acids Res., 44, 2016
|
|
2OIF
 
 | |
5DAI
 
 | |
2PWO
 
 | | Crystal Structure of HIV-1 CA146 A92E Psuedo Cell | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol) | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
2PWM
 
 | | Crystal Structure of HIV-1 CA146 A92E real cell | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
2PXR
 
 | | Crystal Structure of HIV-1 CA146 in the Presence of CAP-1 | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), ZINC ION | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
2QBV
 
 | | Crystal Structure of Intracellular Chorismate Mutase from Mycobacterium Tuberculosis | | Descriptor: | CHORISMATE MUTASE | | Authors: | Ladner, J.E, Reddy, P.T, Kim, S.K, Reddy, S.-K, Nelson, B.C. | | Deposit date: | 2007-06-18 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A comparative biochemical and structural analysis of the intracellular chorismate mutase (Rv0948c) from Mycobacterium tuberculosis H(37)R(v) and the secreted chorismate mutase (y2828) from Yersinia pestis.
Febs J., 275, 2008
|
|
202D
 
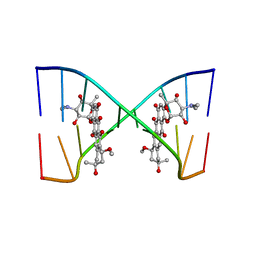 | |
3E2Z
 
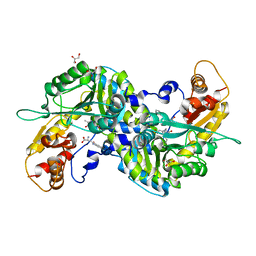 | | Crystal structure of mouse kynurenine aminotransferase III in complex with kynurenine | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLYCEROL, ... | | Authors: | Han, Q, Robinson, R, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2008-08-06 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Correction for Han et al., "Biochemical and Structural Properties of Mouse Kynurenine Aminotransferase III".
Mol. Cell. Biol., 38, 2018
|
|
1OMO
 
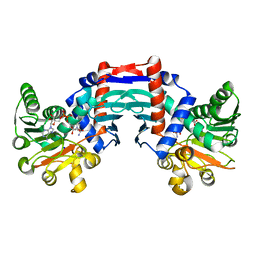 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
3E2F
 
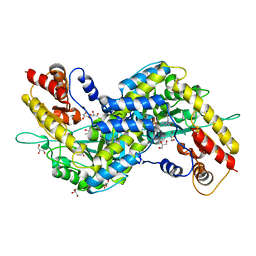 | | Crystal structure of mouse kynurenine aminotransferase III, PLP-bound form | | Descriptor: | GLYCEROL, Kynurenine-oxoglutarate transaminase 3 | | Authors: | Han, Q, Robinson, R, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2008-08-05 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Correction for Han et al., "Biochemical and Structural Properties of Mouse Kynurenine Aminotransferase III".
Mol. Cell. Biol., 38, 2018
|
|
4IOY
 
 | |
3T5S
 
 | |
3N0Y
 
 | | Adenylate cyclase class IV with active site ligand APC | | Descriptor: | Adenylate cyclase 2, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
3N10
 
 | | Product complex of adenylate cyclase class IV | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Adenylate cyclase 2, MANGANESE (II) ION, ... | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
3PSJ
 
 | |
3PSI
 
 | |
3PSF
 
 | |
3E2Y
 
 | | Crystal structure of mouse kynurenine aminotransferase III in complex with glutamine | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLUTAMINE, GLYCEROL, ... | | Authors: | Han, Q, Robinson, R, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2008-08-06 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Correction for Han et al., "Biochemical and Structural Properties of Mouse Kynurenine Aminotransferase III".
Mol. Cell. Biol., 38, 2018
|
|
3N0Z
 
 | |
3PSK
 
 | |
