7UEA
 
 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO1) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
3QTF
 
 | | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity | | Descriptor: | (6S)-6,15,15,18-tetramethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-8,12-(metheno)[1,4,9]triazacyclotetradecino[9,8-a]indole-9-carboxamide, DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, McBean, J.L, Dushin, R.G, Nittoli, T, Ingalls, C, Sutherland, A.G, Sonye, J.P, Eid, C.N, Golas, J, Liu, H, Boschelli, F, Hu, Y, Vogan, E.M, Levin, J.I. | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5703 Å) | | Cite: | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7JST
 
 | | Crystal structure of SARS-CoV-2 3CL in apo form | | Descriptor: | 3C-like proteinase, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT0
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with MAC5576 | | Descriptor: | 3C-like proteinase, PHOSPHATE ION, thiophene-2-carbaldehyde | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JW8
 
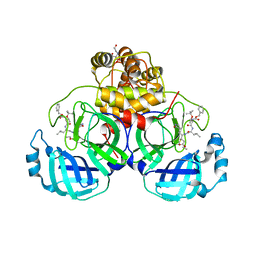 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 in space group P1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-25 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JSU
 
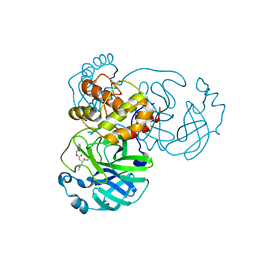 | | Crystal structure of SARS-CoV-2 3CL protease in complex with GC376 | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT7
 
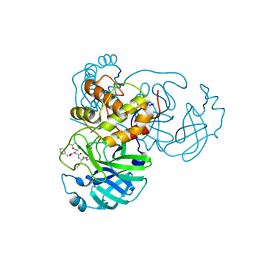 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
3ZFV
 
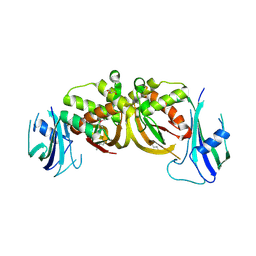 | | Crystal structure of an archaeal CRISPR-associated Cas6 nuclease | | Descriptor: | CRISPR-ASSOCIATED ENDORIBONUCLEASE CAS6 1, GLYCEROL | | Authors: | Reeks, J, Liu, H, White, M.F, Naismith, J.H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Dimeric Crenarchaeal Cas6 Enzyme with an Atypical Active Site for Crispr RNA Processing
Biochem.J., 452, 2013
|
|
7JMO
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-04 heavy chain, COVA2-04 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
7CRJ
 
 | | Dark State Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JVB
 
 | | Crystal structure of the SARS-CoV-2 spike receptor-binding domain (RBD) with nanobody Nb20 | | Descriptor: | CACODYLATE ION, Nanobody Nb20, Spike protein S1 | | Authors: | Xiang, Y, Xiao, Z, Liu, H, Sang, Z, Schneidman-Duhovny, D, Zhang, C, Shi, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.287 Å) | | Cite: | Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2.
Science, 370, 2020
|
|
3B3Q
 
 | | Crystal structure of a synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, X, Liu, H, Shim, A, Focia, P, He, X. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions.
Nat.Struct.Mol.Biol., 15, 2008
|
|
7JMP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody COVA2-39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-39 heavy chain, COVA2-39 light chain, ... | | Authors: | Wu, N.C, Yuan, M, Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Cell Rep, 33, 2020
|
|
7CRI
 
 | | 1 ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8FT4
 
 | | Multicrystal structure of Na+, leucine-bound LeuT determined at 5 keV | | Descriptor: | CHLORIDE ION, LEUCINE, Na(+):neurotransmitter symporter (Snf family), ... | | Authors: | Karasawa, A, Liu, H, Quick, M, Hendrickson, A.H, Liu, Q. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystallographic characterization of sodium ions in a bacterial leucine/sodium symporter
To be Published
|
|
8FT5
 
 | | Crystal structure of LeuT soaked with Crown-5 | | Descriptor: | CHLORIDE ION, LEUCINE, Na(+):neurotransmitter symporter (Snf family), ... | | Authors: | Karasawa, A, Liu, H, Quick, M, Hendrickson, A.H, Liu, Q. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystallographic characterization of sodium ions in a bacterial leucine/sodium symporter
To be Published
|
|
4AVF
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE | | Authors: | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-05-25 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3ZIF
 
 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
7CRL
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 50 ps after light activation | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRY
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (6.49 mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4A3P
 
 | | Structure of USP15 DUSP-UBL deletion mutant | | Descriptor: | ACETATE ION, IODIDE ION, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
